Toward Integrative Omics
Cancer is incredibly complex, posing enormous challenges beyond the biological field. Taking a multi-omic approach can help us make sense of this diverse set of diseases – and, ultimately, allow us to better understand ourselves as human beings.
I started researching colorectal cancer for multiple reasons, but a significant part of my interest was triggered by grief; a member of my immediate family died as a result of metastatic colorectal cancer, despite having the access to the best medical care. I wanted to understand more of what had happened and why.
Reading about colorectal cancer, it was apparent that while the genomics and transcriptomics of the disease had been well studied, the proteomic changes that accompany the disease were not as well understood. I believe the prejudice is related to the tools that were/are available to tackle the problem. After realizing how much remained to be done in the field of cancer proteomics, I decided to devote my career to studying the molecular changes that underwrite colorectal cancer. The more I work in this field, the more I recognize how truly deep understanding – from genotype to phenotype – is the only way we can tackle cancer.
The caterpillar and the butterfly
Multi-omics approaches attempt to make sense of the genome, the transcriptome, the proteome, and the metabolome all together. If you look back to the articles that were written around 2000 (and the publication of the human genome) you will find a glimpse of what we could achieve with this information. With a greater understanding of our genes, transcripts, proteins, and metabolites, we can better understand how the ‘blueprint’ corresponds to reality.
The classic example that I give to my students is the caterpillar and the butterfly. Both have the same genome, but the phenotype of the two animals is shockingly different. That striking physical difference is the result of the transcriptome, the proteome and the metabolome at work.
Multi-omics has of course been gaining traction for the last decade. The major developments that have brought it to the forefront are: i) the completion of the Human Genome Project and ii) the development of high-throughput methods to analyze the transcriptome (first microarrays, later next gen sequencing) and the proteome (mass spectrometry). Multi-omics studies are now everywhere. I would bet that for any major disease there are several manuscripts characterizing the genome, transcriptome, proteome, and/or metabolome of healthy versus diseased tissues. Similarly, it is now routine for the chemical characterization of any organism to start with the sequencing of the genome. When I last checked (April, 2016), the NCBI genome archives held over 75,000 genome sequences, and many of those species will have also been analyzed for transcriptomic and proteomic contents.
In cancer, chemical analysis is highly complex because you are dealing with very different types of molecules that appear at different points in time and in space. For example, a specific transcript or protein may only be needed at certain points in the lifecycle of the organism. If it is only synthesized in a few copies for a short window of time, it can be extremely difficult to measure. Again, I refer to the example of the caterpillar and the butterfly.
Another enormous challenge is the incredible dynamic range of the molecules. Some molecules are produced abundantly at all times, making it hard to see around them. Albumin is the classic example; it makes up more that half the protein content of human blood. What that effectively means is that researchers trying to analyze human blood for other trace level proteins must first deplete albumin before they can conduct any other analyses to see the lower abundant “more interesting” stuff. Separation is the key.
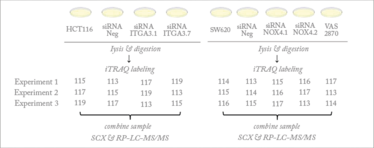
Figure 1. Workflow of experimental design. In brief i) HCT116 cells were silenced with siITGA3 siRNAs ii) NOX4 was inhibited in SW620 cells with siRNAs or VAS2870. Samples were analyzed with a nanoAcquity UPLC (Waters) coupled to a Q Exactive mass spectrometer (Thermo Scientific). Figure taken from reference 1.
Understanding colorectal cancer
When I was a postdoctoral researcher at the National Cancer Institute, almost every member of my lab had lost a family member to cancer. Most of the students who walk into my office tell me they are there because they want to contribute to cancer research. It is a complex problem that affects so many people. From a molecular perspective, it is both fascinating and incredibly motivating. I am hopeful that with greater understanding, we can do a much better job of treating colorectal and other cancers.
Colorectal cancer is a good research target for several reasons. First, it follows a sequential path of genomic instability – more so than other soft epithelial cancers. In colorectal cancer, there is a common pattern of mutations and genomic instability that is observed in approximately two thirds of all colorectal cancer patients. We and others have hypothesized that this similar pattern of genomic instability would result in conserved patterns of proteomic changes – and we are still investigating this phenomenon. Second, though it is one of the most common types of cancer, colorectal cancer is not as well studied as some other cancers. I wonder if the functions of the organs involved result in people being less interested in this disease – unless they have a personal connection. Finally, like many other cancers, colorectal cancer is linked to obesity, meaning that it has the potential to be an increasing health burden in the future.
Driven by mutations
Like many soft epithelial cancers, colorectal cancer starts with a few driver mutations – that is to say, a few mutations that push the cancer along. In fact, there are five critical genes – Pi3K, APC, p53, TGFB, KRAS – that are part of several pathways and have been causally linked to changes in the genome.
Colorectal cancer cells frequently show gross changes in the genome – amplifications and deletions of entire chromosomes are common. And it has been shown that these genomic changes directly correlate with changes to the transcriptome. However, the correlation with the proteome is much less clear. In some of our recent work, we have demonstrated that the amplifications in the genome, while resulting in upregulation of transcripts, do not necessarily result in higher corresponding protein abundance.
Our current understanding of the disease reflects the tools that we employ to detect cancer chemically. For example, to examine changes in the genome, either spectral karyotyping or comparative genomic hybridization are effective analysis strategies. Changes to the transcripts can be assessed by many different high throughput strategies. Microarray analysis is commonly used to survey the expression levels of thousands of transcripts, but are increasingly being replaced by more global next-generation sequencing methods, such as exome and RNA sequencing.
Fundamentally, cancer is a single term used to describe a huge range of diseases. And though the chemical component is very important and dictates the behavior, it’s the phenotype we care the most about at the end of the day. The final definition of whether something is cancer or not is defined by how the cell behaves. Does it grow, proliferate, spread, and metastasize? Although chemicals enable these processes, it is the processes themselves that define the cancer.
Pushing proteomic knowledge
We are examining how protein expression in colorectal cancer differs from normal colon tissue from many different angles. And we are also considering how these expression patterns change over time – as the disease progresses. We use 3D cell cultures to examine spatial differences in expression patterns in tumor mimics.
Our hypothesis is that the genomic changes that are so evident and pervasive in the colorectal cancer genome also play out in the colorectal cancer proteome. As proteins are the action molecules in the cell, they are the best chance we have to develop rational strategies to turn off a cancer-associated signaling pathway.
As we are looking at protein expression, we primarily use mass spectrometry – applying different platforms based on the specific question we are asking. For example, when we are examining the global differences in protein expression level between a normal and a cancer sample, we use quantitative labels and nLC-MS/MS to perform a quantitative comparison (1) – see Figures 1 and 2. When we are examining the differences in spatial distribution in a 3D sample, we employ either imaging mass spectrometry or serial trypsinization to harvest sequential concentric rings of cells for nLC-MS/MS.
We are also interested in gaining a better understanding of how to make treatments more effective. To that end, we’ve developed a powerful imaging approach to visualize drug penetration in tumor mimics, which allows us to see how and where a drug is metabolized. Our approach has been adopted by a couple of European pharmaceutical companies and I hope it will also be implemented by US companies. I believe it could help get more therapies on the market quicker.
We have another project in the lab where we are examining the molecular changes that occur with fasting, also known as caloric restriction. That work has led to some tantalizing evidence that fasting can improve the efficacy of chemotherapies. Now, we are trying to figure out why that is and how it could be implemented clinically.
We’ve also had some extremely rewarding results in the transcriptomics space (see above: The power of transcriptomics and collaboration).
Finally, we are striving to gain a better understanding of why metastasis occurs. The vast majority of cancer deaths result from cells spreading throughout the body. The critical step in the process is the ability of the cells to insert themselves into the secondary location. We are working with Pinar Zorlutuna, a bioengineer, to model a tumor in proximity to a potential secondary site. We have designed the system so that we can manipulate both the chemical and physical stresses. We then evaluate whether the cell succeeds in metastasizing and also evaluate the chemical environment that facilitates or hinders metastasis. I would be delighted if we can decipher a combination of physical and chemical properties that promote – or better yet reject – a metastatic cell. Such information would be incredibly valuable. Thus, five years from now, I hope that we will be applying this knowledge to make potential secondary sites less hospitable for a metastasis.
Cancer is an incredibly complex disease. You can’t effectively treat a disease if you don’t understand it. Our current methods to treat it – radiation, chemotherapy and surgery – are blunt measures. Those in the field share the same hope that with better understanding of the pathways, we can improve diagnosis and therapy.
The power of transcriptomics and collaboration
One of our most striking results comes from a transcriptomics study. We have been working with Steven Buechler, a statistician here at Notre Dame. Steve performs bioinformatics analyses and, a few years ago, he noticed a striking trend in some of the published colorectal cancer microarray studies. In his analyses, he showed that the gene expression patterns were extremely distinct on the right versus the left side of the colon. The colon is a large organ and initially develops in different parts of the embryo, resulting in differential gene expression patterns. The right side of the colon includes the ascending and transverse segments, while the left colon includes the descending colon to the rectum. The two sides of the colon are very distinct; polyp formation differs significantly between the right and the left.
Though it was known that the gene expression patterns between the right and left sides of colon cancer were distinct, the result had never been used clinically. Steve’s lab – in collaboration with my lab – discovered that the gene expressions on the two sides were also prognostic of relapse. We identified a panel of five genes on each side of the colon that can be used to predict whether a patient will have a relapse in the next five years. We’ve been working together over the last few years to validate the expression of these genes in numerous samples (both cell lines and primary tissues) and we hope to translate this information into a clinically actionable test. We are in the process of publishing these results and patenting the tests, so I can’t say more at this moment, but I am extremely excited about this work. It’s a project that could make a real difference to people’s lives.
We couldn’t do the project without Steve Buechler; he brings the statistical expertise and we have the bench-top know-how. The project only works when we both work together. In fact, many of the current projects in the labs are only made possible through collaboration with other research groups.
Omics in the Literature
What does analysis of the last 15 years of literature on the different omics tell us about the growing importance of the field, and the move towards a more integrated approach?
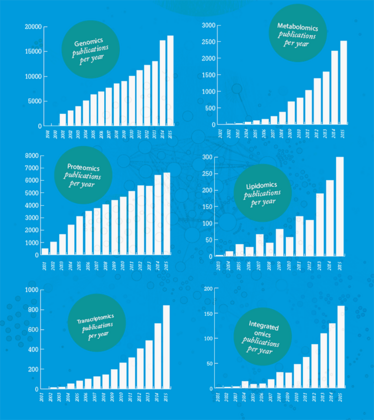
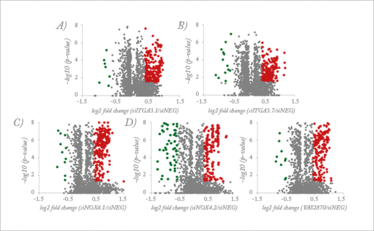
Figure 2. Volcano plots displaying protein expression changes that are statistically significant. (A, B) Proteins changed in expression with ITGA3 silencing. (C−E) Protein altered in expression with NOX4 gene silencing or chemical inhibition. Green data points = downregulated proteins; red data points = upregulated proteins. Figure taken from Reference 1.
Enabled by technology
It’s clear that advances in next-gen sequencing are driving genomics. But for those of use placing an emphasis on proteomics, the most important technical advances are improvements in mass spectrometers. About ten years ago, the Orbitrap mass analyzer hit the market, making high-resolution instrumentation less expensive. Prior to that point, the only available high-resolution instruments were ion cyclotron resonance MS systems, which were prohibitively expensive for most labs. And though Orbitrap technology is expensive, it is relatively more affordable and has enabled global proteomic analyses in a way that wasn’t possible a few years ago.
To truly enable advances in the field, mass spectrometry must be paired with really smart data analysis. So I’d like to acknowledge the incredible importance of database searching algorithms, such as MASCOT and SEQUEST. Using such search tools, we can rapidly identify thousands of peptides, and thus proteins, from a complex mixture.
The development of these tools was really seminal for the field of proteomics – a fact that becomes more evident when you consider the current state of metabolomics. In metabolomics research, the separations and mass spectrometric analyses are similar to proteomics research. However, the databases and the search algorithms are not yet mature. The current standard practice to confirm identification is to test your compound of interest against a known standard, which is expensive, labor intensive and low throughput. As a result, while the field is growing, there isn’t a widespread consensus on how to identify features. I anticipate that within the next few years, someone will develop an approach that enables rapid confirmation of mass spectrometric metabolite datasets, which would be transformative for the field, gain great traction and have a huge scientific impact.
Our omics-driven future
Going back to multi-omics, there is an excellent article from Shelia Jasanoff, in which she compares the human genome to the US Constitution: “Like the Constitution of the United States, the human genome turned out to be a sparse document, containing fewer genes than expected. This means that, as with the Constitution, the genome’s meanings will evolve over time, as scientists, lawmakers, and [the public] make sense of the fixed elements of the sequence in relation to the variables and unknowns in the surrounding environment.” She also addresses some of the criticism that has been leveled at the Human Genome Project and the fact that it has not resulted in fast medical breakthroughs: “A decade is not nearly enough time to measure the impact of a scientific revolution [...] It is too soon to tell whether cures for genetic disease were oversold [...] What matters is that we found a powerful new way to represent human identity, and the moral implications of that re-representation are just beginning to unfold.”
I like these analogies. I think the problems we are trying to answer are incredibly complex and it is unrealistic to think that huge sweeping medical changes will result immediately. That being said, there are already some medical changes occurring. Just in the last couple years, it has become possible for pregnant women to learn about the genomic status of their fetus through circulating fetal (cf) DNA sampled from a blood draw. That is an enormous advance and I anticipate that within a few years a range of tests will be available on cfDNA and other valuable samples. Similarly, another area of research that I think is on the cusp on making a breakthrough scientifically is the analysis of circulating tumor cells.
Tumors shed cells into the bloodstream and researchers are making great strides in their ability to enrich for these cells and perform omics analyses on them. Success in this arena would have a huge impact on cancer diagnoses in the next few years. Both of these developments fall under the umbrella of personalized diagnostics. And I anticipate that we will see many more of these important developments in the near future.
- KM Bauer et al, “Proteomic and functional investigation of the colon cancer relapse-associated genes NOX4 and ITGA3”, J Proteome Res. 13(11), 4910-8 (2014). DOI: 10.1021/pr500557n.
- S Jasanoff “Genome-sequencing anniversary. A living constitution”, Science 331(6019), 872. DOI: 10.1126/science.1203467
Amanda Hummon is Huisking Foundation Inc. Associate Professor at University of Notre Dame, Indianapolis, USA. The interests of the Hummon Research Group lie at the intersection of analytical chemistry and chemical biology, with a focus on cancer biology. They develop high-throughput methods to evaluate the proteome and transcriptome in cancer cells.