An international team has unveiled new mechanisms for delivering messenger RNA (mRNA) to bacterial ribosomes, using cryogenic electron microscopy (cryo-EM), single-molecule kinetic analysis, and structural proteomics to offer detailed insights into the molecular process of gene expression.
One of the mechanisms involves the Shine-Dalgarno (SD) motif, a conserved RNA sequence that helps align the ribosome with mRNA for decoding. Additionally, ribosomal protein S1 and RNA polymerase, which produces mRNA, contribute to guiding the mRNA to the ribosome.
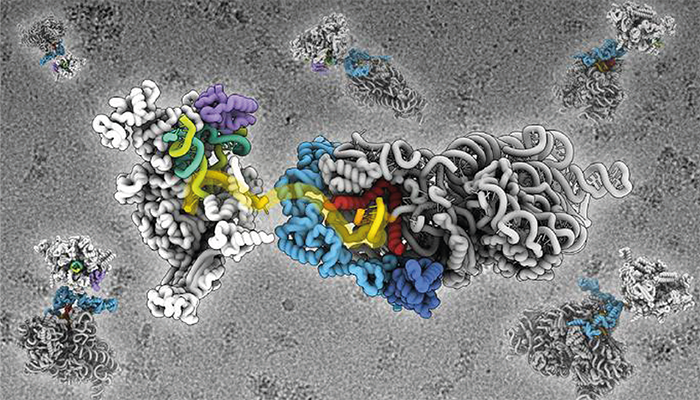
Credit: John Innes Centre
Cryo-EM was used to create atomic-level models of ribosome-mRNA assemblies; by isolating ribosomes and assembling them with other components in states that mimic mRNA encounters, researchers visualized a pathway on the ribosome surface that directs mRNA toward the decoding site. Single-molecule kinetic analysis was used to measure the dynamic interactions between mRNA and ribosomes, providing real-time insights into how these molecular components find and interact with one another. Structural proteomics further confirmed the connections between ribosomal proteins, RNA polymerase, and mRNA within bacterial cells, corroborating the cryo-EM findings.
“The surprising finding is that there really is a mechanism that has evolved to help mRNAs be delivered to the ribosome for translation,” said Michael Webster, a group leader at the John Innes Centre and co-author of the study, in a press release. “I was surprised how clearly our structural models show that the bacterial ribosome can make a path for the incoming mRNA.”
The mechanisms described may also be relevant to chloroplasts in plants, which share evolutionary origins with bacteria. Understanding these processes could inform future research into photosynthetic protein production and potentially contribute to developing crops better suited to climate challenges.
The collaborative study involved expertise from the John Innes Centre, the IGBMC in Strasbourg, the University of Michigan, and the Technische Universität in Berlin.
