Introduction
Etanercept has been widely used in the treatment of plaque psoriasis, psoriatic arthritis, ankylosing spondylitis, and rheumatoid arthritis for over 20 years.1, 2 It works as a tumor necrosis factor (TNF) antagonist by binding to TNF molecules and preventing them from binding to the cell-bound receptors—which could otherwise lead to autoimmune disease when de-regulated TNF production occurs.3 Etanercept is a heavily glycosylated fusion protein consisting of the extracellular ligand-binding domain of human TNF receptor 2 (TNFR2) and the Fc domain of human IgG1.4 There are 2 N-glycosylation sites and 13 potential O-glycosylation sites on the TNFR2 domain and one N-glycosylation site on the Fc domain.4 The glycosylation alone represents approximately 30% of the total molecular weight of etanercept.5 The fusion protein is also a homodimer where disulfide bonds connect the two monomers. These properties combined make the protein structure very complex and highly heterogeneous. The physicochemical characterization of such molecules is very challenging.
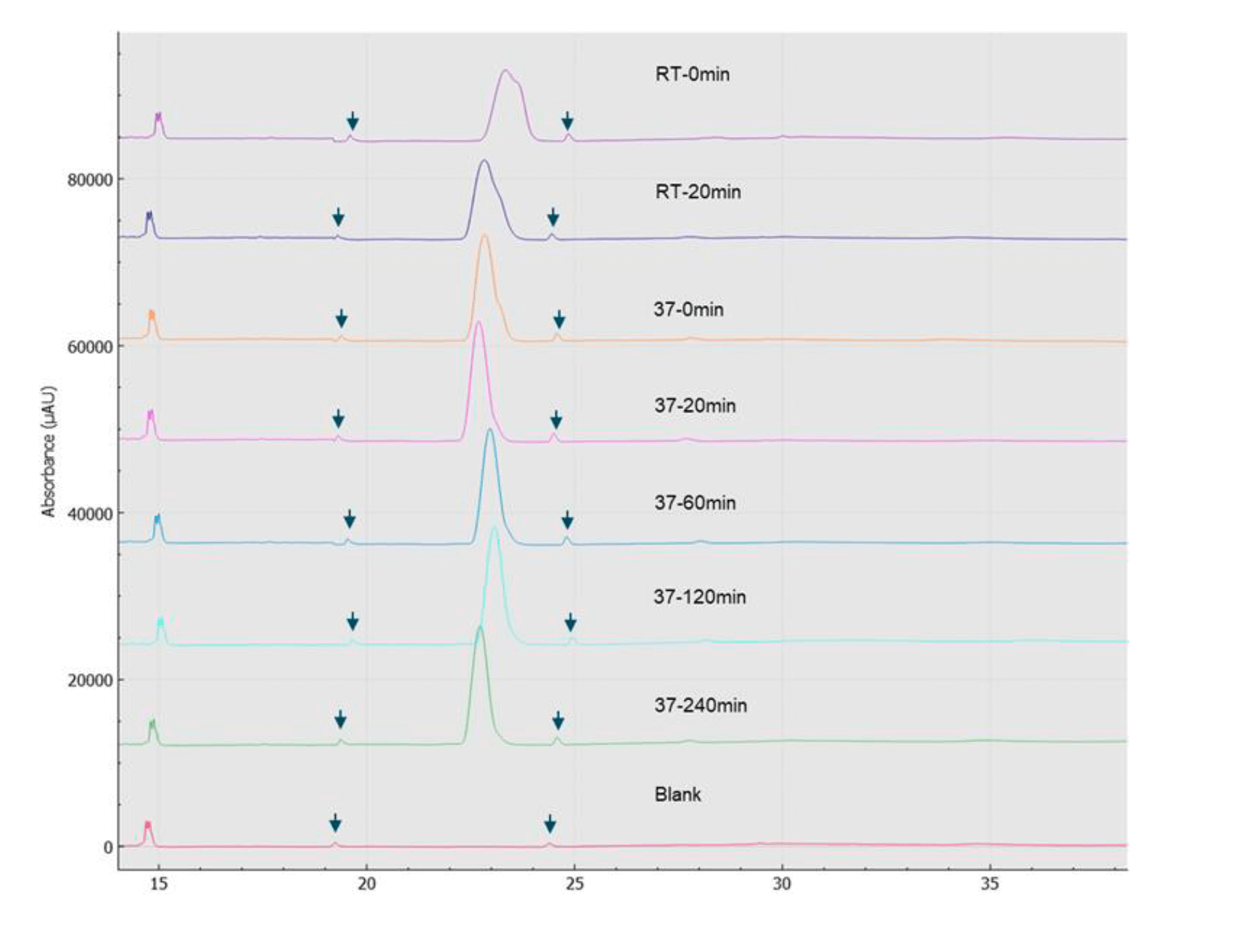
Capillary electrophoresis-sodium dodecyl sulfate (CE-SDS) separates proteins based on size. It has been used in applications such as protein characterization, purity assessment, stability testing and lot release.6 It is one of the most powerful separation techniques in the analytical chemistry toolbox because it offers a high speed of analysis, high resolution, and quantification. It is robust and reproducible and amenable to automation. In this work, the BioPhase 8800 system (Figure 1) was used to analyze etanercept by capillary electrophoresis-sodium dodecyl sulfate (CE-SDS). The automated capillary electrophoresis instrument offers fast separation times and proven high reproducibility.6, 7 When faced with the challenge of analyzing complex proteins under different conditions, performing CE-SDS analysis one separation at a time can be time-consuming. The multi-capillary capability of the BioPhase 8800 system allowed for the simultaneous analysis of etanercept under different deglycosylation reaction conditions, deglycosylation time-course, and assessing the consistency of deglycosylation reactions in other lots, accelerating methods development for protein analysis.





