In contrast to conventional LC columns that contain randomly packed beads as their stationary phase, microchip-based pillar array chromatography columns have a separation bed of perfectly ordered and freestanding pillars obtained by lithographic etching of a silicon wafer. The regular mobile phase flow pattern through these microchip pillar array columns adds very little dispersion to the overall separation, resulting in better peak resolution, sharper elution and increased sensitivity. The freestanding nature of the pillars also leads to much lower back pressure buildup, and makes it possible to operate longer columns.
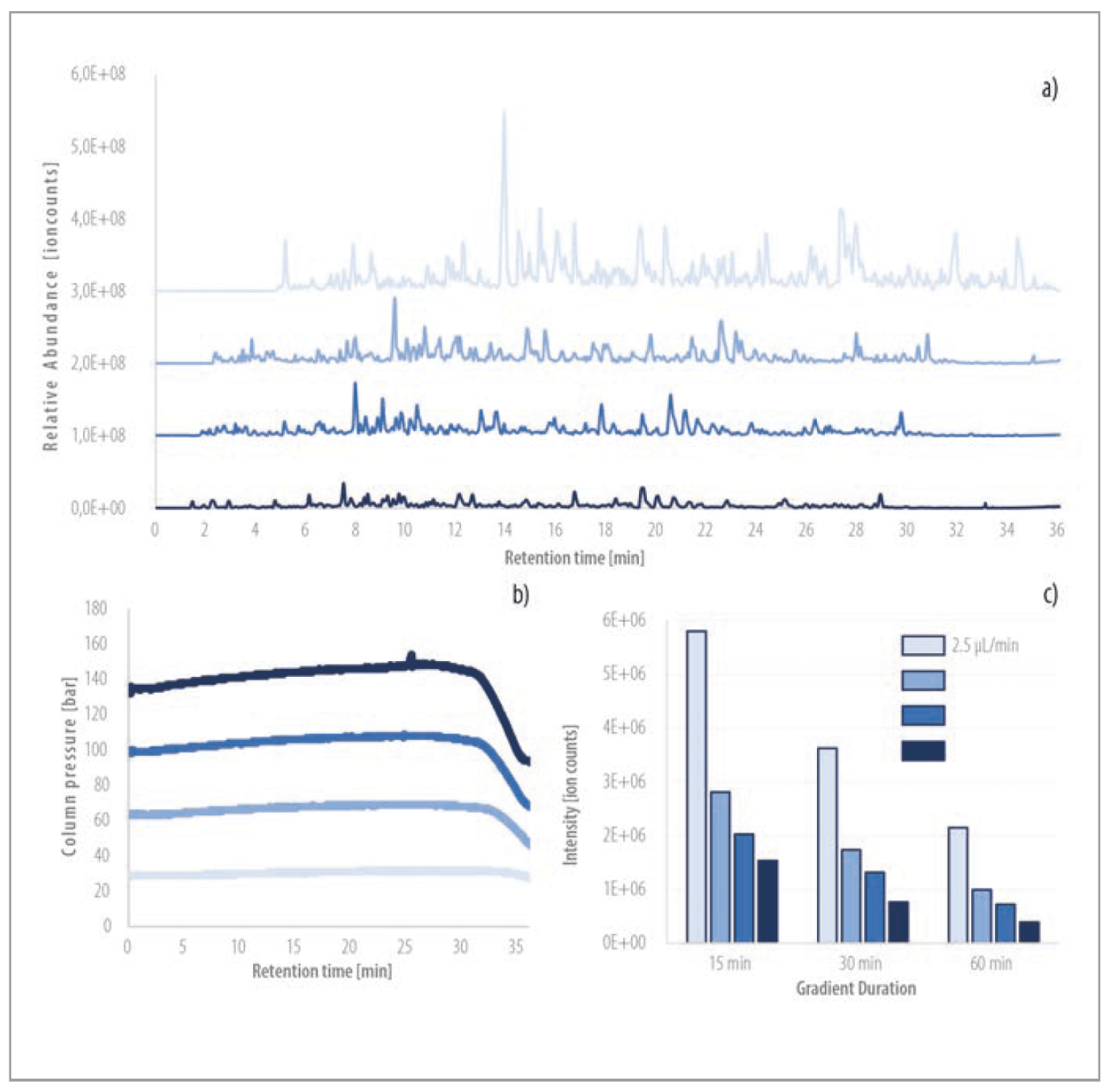
As a consequence of the low column backpressure inherent to μPAC™ column backbones, chromatographic separation can be performed over a wide range of flow rates (1–15 μL/min). MS base peak chromatograms of 30-min gradient separations (500 ng HeLa protein digest standard) are described for four different flow rates (2.5, 5, 7.5 and 10 μL/min). Whereas the first eluting peptide peaks are observed at a retention time of 5 min at 2.5 μL/min, operating the column at 10 μL/min allows reducing the column void time down to 1 min, which can result in a significant increase of sample throughput for bottom-up proteomics experiments. Besides a high flow rate flexibility and low column backpressure, excellent peptide peak shapes were obtained with the μPAC™ capLC column. Based on 15 reference peptides from the Pierce™ Retention time calibration (PRTC) mixture, average peak widths measured at half maximum (FWHM) between 0.10 and 0.11 min were achieved for the 30-min gradient separations at all flow rates.
When comparing the μPAC™ capLC column to a packed bed alternative, striking differences in retention time stability were observed. Whereas an average coefficient of variation on retention times (including all 15 PRTC peptides) of 0.73 percent was observed for the packed bed column, variation in retention time was reduced almost three-fold (down to 0.26 percent CV) by working with the μPAC™ column. This highlights again the positioning of the μPAC™ capLC column as a robust and reliable alternative to the traditional packed bed capillary flow columns frequently used in high-throughput proteomics research.
Considering the overall number of identified protein and peptide groups, it becomes clear that the μPAC™ capLC column outperforms the traditional packed bed column. For a 30-min gradient at a flow rate of 7.5 μL/min, a relative gain of more than 20 percent at the protein level and more than 40 percent at the peptide level was found for the μPAC™ capLC column. But also for the other applied gradient times (15–60 min) and flow rates (2.5–10 μL/min), the PharmaFluidics μPAC™ capLC column outperforms conventional state-of-the-art columns, resulting in a higher proteome coverage (both at protein and peptide level).
Jeff Op De Beeck, Geert Van Raemdonck, Bo Claerebout, Kurt Van Mol, Natalie Van Landuyt and Paul Jacobs