Chemical structure verification by NMR is one of the most fundamental, and yet challenging practices in synthetic chemistry. With the overwhelming expansion in the volume of data and size of compound libraries, automated structure verification using NMR data is becoming an invaluable tool for timely and unambiguous characterization in the synthetic workflow. ACD/NMR Workbook Suite is the only commercially available software that can perform structural verification at different levels for added confidence and flexibility in the analysis.
In the simplest workflow, referred to as Single Structure Verification (SSV), a proposed structure selected based on the knowledge and expected chemistry of the sample is submitted along with the experimental NMR data. A single 1D or 2D NMR spectrum,1 or a combination of 1D and 2D spectra is required for this method. The software evaluates the match between the proposed structure and the datasets in the NMR project and reports a match factor (MF). While it can be confidently confirmed when a proposed structure fails this “NMR filter”, without alternative structures presented as potential “better fits”, there is an increased chance for false positives.
In order to confidently reduce the false positive rate, in the next protocol referred to as Combined and Concurrent Verification (CCV), the advanced algorithms concurrently verify the fit between a specified number of generated isomers with the proposed structure.2 The MF rankings will indicate if any of these alternative structures are more consistent with the NMR data. However, since this protocol matches the experimental data against a selected number of isomers defined by the user or an algorithm, the initial bias from the chemist may still affect the results.
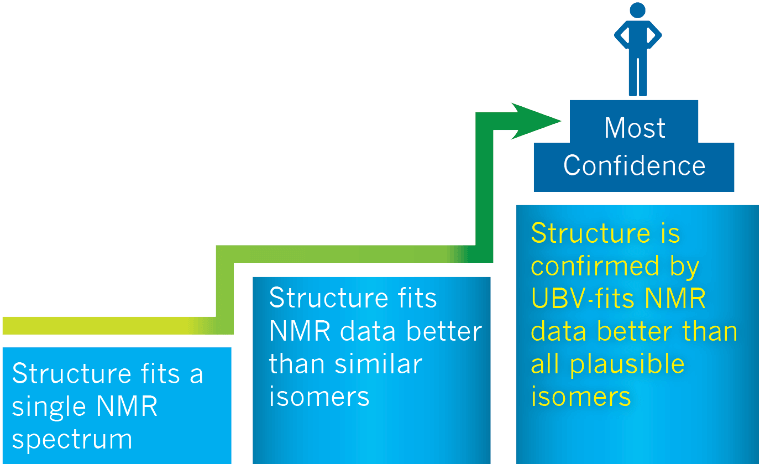 Figure 1. ACD/NMR Workbook Suite performs structural verification at different levels
Figure 1. ACD/NMR Workbook Suite performs structural verification at different levels




