Bernd Kammerer heads the metabolomics facility at the Center for Biosystems Analysis (ZBSA), University of Freiburg, Germany, the purpose of which is to identify and quantify the full range of metabolites in biological systems. Bernd has broad scientific experience in metabolite and metabolome analysis, particularly with MS and NMR methods, and is familiar with bioinformatic methods for cluster analysis and metabolomic data mining. What are the specific analytical needs of metabolomics? Well, the complexity and high dynamic range of metabolite concentrations pose tough challenges for qualitative and quantitative analysis. Typically, the target compounds are small molecules with masses ranging between 100 and 1000 Da. These are studies in a wide variety of biological matrices, such as cell culture samples, urine and blood. On the one hand, you need precise MS analysis; on the other hand, a highly effective chromatographic separation is indispensible. How has your view of 2D-LC changed over the last decade? Ten years ago, 2D-LC was accomplished mainly by coupling the first and second dimensions offline. This provided fantastic separation power but it was very time-consuming, something that has been solved by modern 2D-LC systems. Today, it is possible to perform comprehensive 2D-LC without loss of time and with fewer potential sources of error. What problem were you addressing when you first considered 2D-LC? A major research project for us is the identification of a metabolic signature for early detection of human breast cancer. To do this, we are analyzing different biological samples with a high degree of complexity. Our target substances – modified nucleosides and ribosyl derivatives – differ only slightly in terms of their chemical structure and, therefore, retention in LC. Consequently, we decided to lift the chromatography to the next level, that is, to two-dimensional separation. Metabolomics generally deals with complex matrices containing several hundred compounds, so separation power is essential. Insufficient separation can lead to the formation of highly reactive radical cations that interact with each other in the ion source (ESI or APCI) before entering the MS, causing ion suppression and artefacts. Since we are often looking to distinguish between two different biological states, it is important to improve the (semi-)quantitative analysis of target compounds, something that has particular relevance to ion suppression. An additional advantage of increased separation power is the possibility of determining previously hidden metabolites. What were your expectations of 2D-LC? Two things really. The structural class we deal with is differentially modified nucleosides, and we expected to achieve a better differentiation between isomers and nucleosides that could hardly be separated at all using a single column. And we were hoping for purification of peaks in the second dimension separation to help limit background noise. What has been your experience so far? We have just started with the new technique and are in the process of method optimization. Choosing the right separation mechanism is the first step of method development. Column dimensions, solvents and elution gradients all offer many possibilities to improve your analysis and different combinations of column materials lead to striking differences in orthogonality, retention and peak capacity. Our 2D-LC can be used as a standalone solution with a diode array detector, but it can also be coupled to different mass spectrometers. This high degree of functionality and flexibility is important for studies of complex biochemical pathways. Currently, we are looking forward to the first comprehensive measurements from a large batch of real life samples. The first results look very promising (see Figure 1).

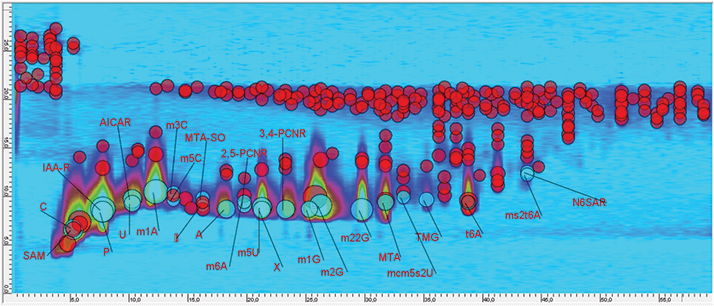
Do you anticipate 2D-LC being adopted more fully in metabolomics? Yes. The importance of comprehensive 2D-LC will grow rapidly because of its universal applicability. I expect that 2D-LC in combination with different ion sources and MS solutions will become essential in applications of analytical chemistry, especially in research that must cope with complex matrices and/or complex analyte spectra. How will you use 2D-LC in the near future? We are planning to combine our 2D-LC solution with different mass spectrometers to address a range of sample and chromatographic challenges. Optimization of the chromatographic methods for the particular challenge at hand is clearly important here. In particular, for the separation of structural isomers, which occur frequently in modified nucleosides, I can see 2D-LC opening up new vistas. Read about the other new kid on the block here.

