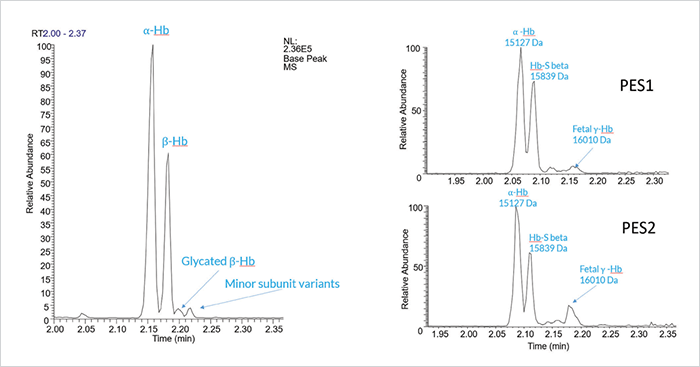
Overview
Whole blood or purified blood cells are often analyzed in an effort to identify specific compounds that may be indicative of disease states. The compounds that are measured can range from small molecule metabolites to larger protein biomarkers.1,2 This necessitates the need for multiple pieces of instrumentation as there are few techniques capable of effectively analyzing both small and large molecules rapidly and simply. In this work we present a microfluidic CE-MS system capable of analyzing large and small molecule biomarkers without the need to significantly alter the instrumentation. The ZipChip® microfluidic CE-MS interface coupled to Thermo mass spectrometers was used to analyze small molecule and protein based biomarkers from a variety of blood samples.

Methods
Sample Preparation and Analysis. Although liquid blood samples could be used, samples for small molecule assays were derived from dried blood spots (DBS). The DBS were created by blotting samples of whole blood onto filter paper and allowing the spots to dry. To simulate samples indicative of disease states, phenylalanine or octanoylcarnitine were spiked into blood samples prior to blotting on the filter paper. 5 mm punches of the DBS were extracted with 250 μL of a MeOH/water/ammonium acetate solution doped with stable isotope internal standards (Cambridge Isotope Laboratories, Inc.). DBS punches were incubated in the extraction solution for 30 min at room T and then filtered with a 0.22 μm Nylon filter to remove particulates. Protein biomarkers were measured from whole blood or isolated erythrocytes by lysing the cells with an ACN/ water/formic acid solution. Samples were filtered with a 0.22 μm Nylon filter to remove particulates.