By visualizing molecular interactions in the cell membrane with a pioneering mass spectrometry (MS) method, researchers at Texas A&M University have shown how proteins are able to recruit a specific lipid microenvironment via allostery (1). A common feedback mechanism in biology, allostery is the regulation or modulation of a biological macromolecule through the binding of an effector ligand to a binding site other than the active site. The Texas researchers have shown that this method of communication extends to lipids. When monitoring individual lipid binding with native ion mobility MS – a technique pioneered by group leader Arthur Laganowsky that preserves non-covalent interactions – the group noted that different lipid pairs exhibit various degrees of allosteric modulation.
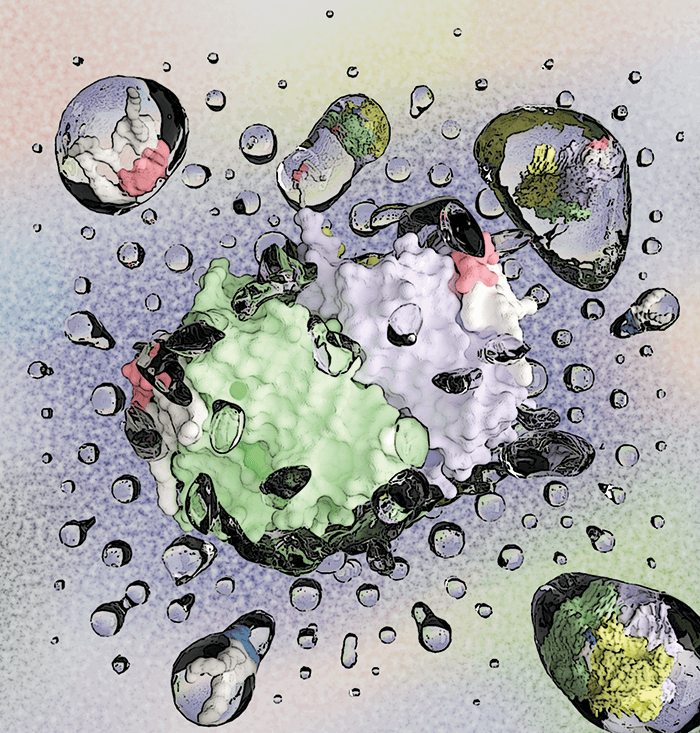
“It is becoming increasingly clear that membrane proteins are exquisitely sensitive to the chemistry of the lipid,” said Arthur Laganowsky in a press release. “Given that lipid composition differs throughout the organs of the body, understanding how the lipid environment in these areas influences protein structure will be critical to opening new possibilities for pharmaceutical drugs designed to affect how these lipids bind with one another.” Lipid–protein interactions are generating increasing interest, as scientists uncover the key role played by lipids in the structure and function of membrane proteins; far from being inert, the lipid microenvironment of the membrane apparently plays an active part in the functioning of the cell.
References
- JW Patrick et al., “Allostery revealed within lipid binding events to membrane proteins”, Proc Natl Acad Sci (2018). Available at: https://bit.ly/2HEZK46. Accessed April 30, 2018.




