In recent years, breath analysis has emerged as a non-invasive and promising tool for disease diagnosis. Hundreds of chemical compounds are encompassed within a breath sample, and volatile organic compounds (VOCs) in particular can be derived from metabolic processes. Measuring VOCs in breath samples provides a snapshot of the physiological status of bodily functions – the composition and abundance reflecting metabolic changes that could be indicative of disease.
Unlike current sampling methodologies, which mainly focus on fecal matter and blood, breath collection provides a non-invasive and convenient method to real-time testing. As a waste product constantly emitted from the body, breath sampling enables researchers to take repeated measurements over a short timeframe with minimal discomfort to the patient.
In 2020, The Analytical Scientist published a feature on making the “breath biopsy” a reality. This article will focus on recent advances in our understanding of how microbiome-produced VOCs in breath add to the picture.

Breath analysis of the gut microbiome
Many studies have demonstrated distinct breath VOC profiles amongst individuals with and without specific diseases. Such VOCs have the potential to distinguish individuals with particular diseases, facilitate diagnosis, and provide a tool for monitoring disease progression and treatment efficacy.
Various analytical approaches can identify VOCs in a breath sample. However, gas chromatography-mass spectrometry (GC-MS) is widely accepted as the gold standard due to its high sensitivity and accuracy. Depending on the goals of the study, additional computational approaches – such as regression analysis – can generate discriminatory tests from breath samples. After identifying molecular features via a mass spectrum in GC-MS analysis, these features are matched to libraries like the National Institute of Standards and Technology (NIST) for tentative identifications.
VOCs produced by gut microbes – such as short-chain fatty acids (SCFA), indole, trimethylamine, and other compounds involved in different metabolic pathways – can also be found in breath samples. The changes in abundance of these compounds in traditional sampling matrices (fecal matter and blood) suggest the involvement of the gut microbiome in gastrointestinal diseases and cardiovascular metabolic diseases (1–4).
The gut microbiome in itself is highly diverse and variable depending on several factors, including lifestyle, age, and overall health. Differences in the community of microbes have also been associated with the development of inflammatory bowel disease (IBD) (5,6) and cancer (7,8).
A study in 2018 demonstrated the impact of the microbiome in Crohn’s disease – a subtype of IBD (9). During active and remission status, individuals provided breath samples for non-targeted VOC analysis and fecal samples for metagenomics analysis. The former approach doesn’t require prior VOC identification, allowing for the discovery of novel biomarkers. Additionally, this approach captures all the compounds reflecting overall physiological changes, decreasing bias, and improving ideology for exploratory studies.
In this particular study, compounds were analyzed with thermal-desorption GC-MS, with wavelets and P-splines applied to reduce background noise and correct baseline respectively. To determine which compounds were the same, the area under each peak of the total ion current (TIC) chromatograms and similarities in retention time were calculated, while the correlation of mass spectra were matched among similar samples. A data matrix with patients and relative concentrations of VOCs were also created for correction analysis.
Though the authors did not validate identified compounds with a chemical standard, the results showed that, regardless of disease status, the tentatively identified SCFAs, acetate, and propionate were significantly correlated with levels of Bifidobacteria and several other microbes in the Firmicutes phylum – all of which were implicated in the development of Crohn’s disease.
Additionally, the abundance of these VOCs and their correlated microbial strains decreased in subjects during their disease state relative to their remission state. This demonstrates how microbial-produced breath VOCs, the gut microbiota, and disease state are implicitly connected.
Gut microbial activity in drug development
With further research into microbiome-produced VOCs in breath, we could see a revolution in disease diagnosis and monitoring for liver disease.
Non-alcoholic fatty acid liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH) are thought to share common physiological causes with alcoholic fatty liver disease (AFLD). Recent studies concluded that microbial dysbiosis may be associated with the development of NAFLD and NASH through overproduction of ethanol by certain bacteria, such as Klebsiella pneumoniae (10,11). In other words, without consuming alcoholic beverages, the bacterial-derived ethanol is high enough to exceed the recommended daily amount.
These findings could have a knock-on effect on the pharmaceutical industry, given that nearly half of prescribed medications have potential interactions with alcohol. As shown in Figure 1 below, there are at least eight classes of drugs in development for NASH treatment – each acting against different relevant targets to alleviate symptoms or slow disease progression. Disregarding gut ethanol production during the development of these treatments could lead to clinical trial failure and reduce the chances of establishing drug efficacy. A diagnostic test for bacterial ethanol production could also help identify individuals at risk of high ethanol exposure prior to clinical visits, enabling better selection of patients for drug trials or treatment.
As ethanol is metabolized by alcohol dehydrogenase in the liver before reaching the peripheral circulation, concentrations of bacterial-derived ethanol in exhaled breath are expected to be less than 20 ppb (parts per billion) – much lower than the current limit of detection of commercially available breathalyzers. Lab-based analytical instruments are capable of detecting breath ethanol in the ppq (parts per quadrillion) range and, with further development, can be translated to handheld sampling devices for at-home testing.
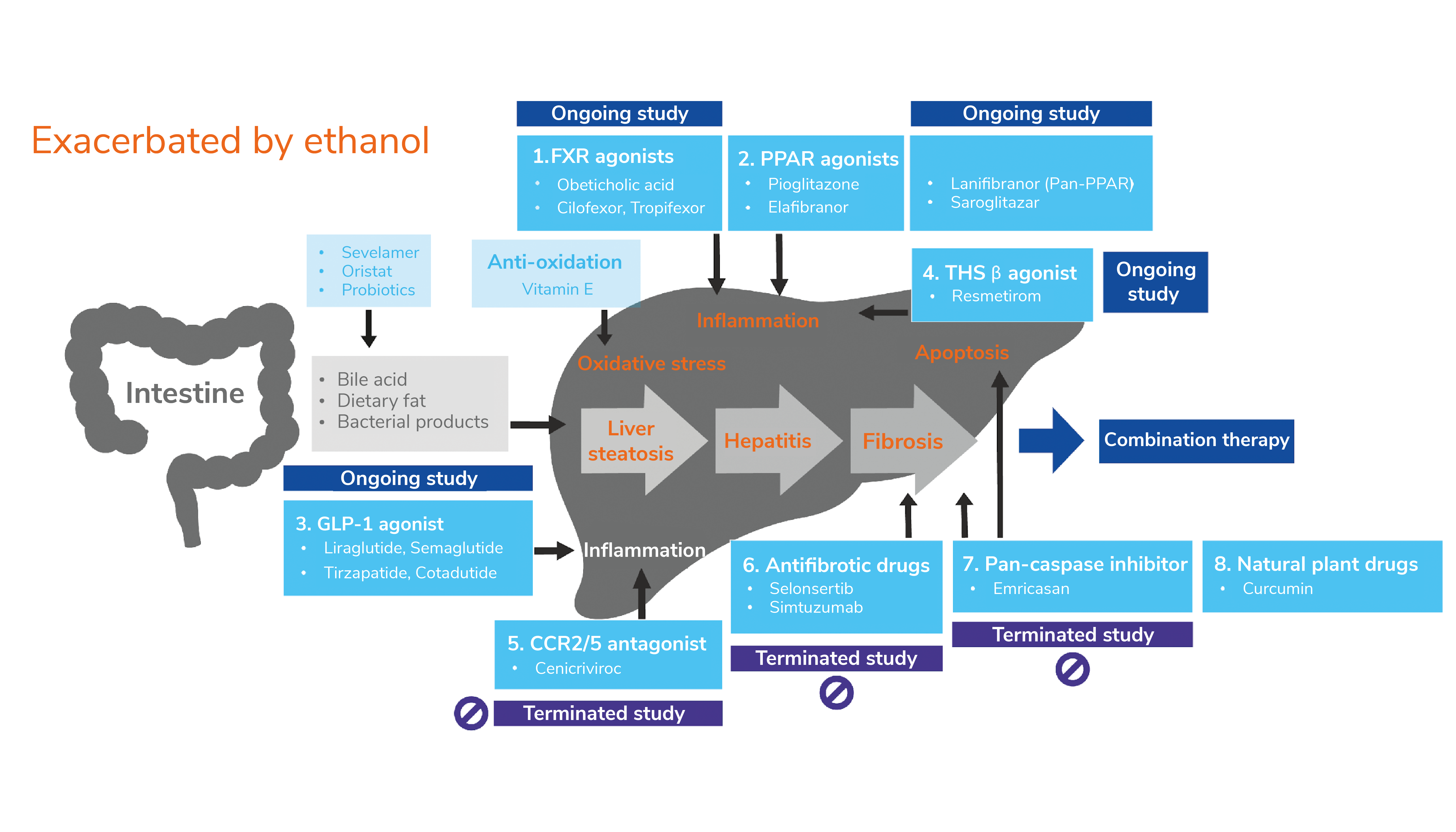
Figure 1. Pharmacological targets for NASH therapy
Fighting antimicrobial resistance
The intersection between breath analysis and the human microbiome does not stop at disease diagnostics and treatment monitoring. Integration of breath and microbiome research can provide further insight into rising antimicrobial resistance concerns – potentially improving clinicians’ ability to reduce this issue alongside other health risks through tailored therapy.
A pilot study hypothesized that the treatment pathway and chosen antibiotic may have varying effects on the patient’s gut microbiome – with changes reflected in breath volatile metabolites (12). Over a 12 month period, the patient’s gut microbiome and breath volatile metabolite data were recorded before, during, and after antibiotic therapy. Breath samples were analyzed through GC-MS, while microbial DNA from stools were analyzed through long and short-read DNA sequencing.
The relationship between breath and gut microbiota in this study was less clear for several reasons, including a lack of full representation of the gut microbiome in fecal matter. Additionally, the study conducted a targeted analysis, which didn’t represent the whole metabolome and could have missed crucial information.
However, an interesting result was found in one of the patients referred to prolonged antimicrobial therapy due to an unusual clinical history, with 11 years of biliary system infection. This patient had consistently high concentrations of all methyl halides, which negatively correlated with three specific gut bacteria. Between pre and post-therapy, a shift in the dominant Bacteroides strain was noted in this patient, along with altered breath metabolite composition.
Though the patient’s health did improve over time, most breath VOCs didn’t return to their baseline values, suggesting that microbial or human metabolism was substantially modified during recovery and/or antimicrobial treatment. The longer-term effects of microbial changes and the VOCs produced through treatment remain unknown – further research into these specific details is required. However, the case report shows the significance of comparing longitudinal pattern changes: understanding the effects of antimicrobial exposure through breath and microbiomes is an important factor in improving personalized medication in the years to come.
The need for breath sampling
Microbial-produced VOCs are typically more abundant than those endogenously produced in a breath sample, but volatile compounds in general are in low abundance in exhaled breath. Therefore, there is a crucial challenge in effectively reducing noise in samples and enhancing the sensitivity of compound detection. Apart from restricting test subjects’ behaviors (such as drinking, eating, and smoking) prior to breath sampling, noise reduction can be achieved by ensuring the compound measured is genuinely “on-breath.”
Once identified and validated, breath biomarkers can be used for a targeted chemical analysis approach, with less concern for background reduction. A validated biomarker or a panel of biomarkers can be leveraged by technology like field asymmetric ion mobility spectrometry (FAIMS) or metal oxide sensors to generate a handheld device for clinical tests. For example, hydrogen-methane sensor-based tests provide rapid, accurate, and cost-effective diagnosis for gastrointestinal conditions, such as small intestinal bacterial overgrowth (SIBO) and carbohydrate malabsorption.
Overall, breath sampling provides researchers a non-invasive and user-friendly approach to obtain real-time data reflecting changes in microbial-produced metabolites. This enables informed decision-making in drug development and the customization of treatment plans. With a vast range of application possibilities, there is a clear demand for an effective breath sampling device to analyze the metabolic activity of the gut microbiome and the screening and diagnosis of associated diseases.
Headshot and Figure - Credit: Supplied by Author
References
- X Cui et al., SciRep, 8, 635 (2018). DOI: 10.1038/s41598-017-18756-2.
- DM Moutsoglou et al., Am. J. Respir. Crit. Care Med, 207, 6 (2023). DOI: 10.1164/rccm.202203-0490OC.
- NC Ward et al., J Hypertens, 40, 8 (2022). DOI: 10.1097/HJH.0000000000003190.
- C Tana et al., Neurogastroenterol Motil, 22, 5 (2010). DOI: 10.1111/j.1365-2982.2009.01427.x.
- DN Frank et al., Proc Natl Acad Sci U S A, 104, 34 (2007). DOI: 10.1073/pnas.0706625104.
- G Le Gall et al., J Proteome Res, 10, 9 (2011). DOI: 10.1021/pr2003598.
- T Jain et al., Front Immunol, 12 (2021). DOI: 10.3389/fimmu.2021.622064.
- S Kim et al., J Chromatogr Sci, 57, 5 (2019). DOI: 10.1093/chromsci/bmz011.
- A Smolinska et al., Anal Chim Acta, 1025 (2018). DOI: 10.1016/j.aca.2018.03.046.
- L Zhu et al., Hepatology, 57, 2 (2013). DOI: 10.1002/hep.26093.
- J Yuan et al., Cell Metab, 30, 6 (2019). DOI: 10.1016/j.cmet.2019.08.018.
- F Shahi et al., Wellcome Open Res, 7 (2022) DOI: 10.12688/wellcomeopenres.17450.3.




