The Analytical Scientist × Thermo Fisher Scientific
Earlier this year, I had the pleasure of delivering the plenary lecture at the 1st International Symposium on Recent Developments in Pesticide Analysis in Prague, Czech Republic (watch the presentation online at: http://tas.txp.to/1115/Mastovska I wanted to provoke discussion, and so decided on a bold (perhaps even intimidating) title: “New and Never-Ending Challenges for Pesticide Routine Testing Laboratories.” Why do the challenges feel never-ending? Firstly, pesticide residue analysis must constantly react to three (ever-changing) compounding factors: large numbers of analytes, low limits of detection, and a diversity of matrices.).

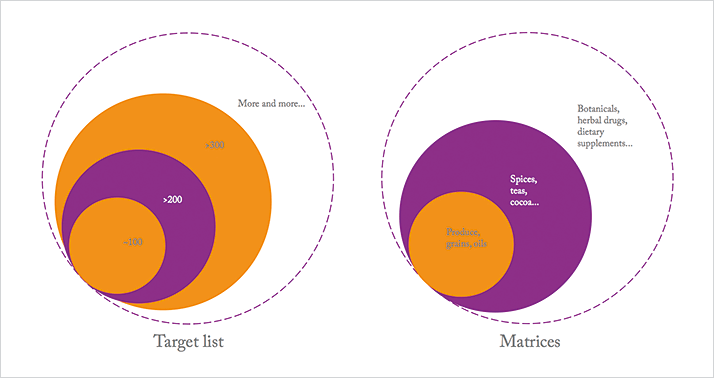
Moreover, the increasingly global nature of trade in the food industry adds to the mix. Wider sourcing of raw materials (and distribution of products), unknown pesticide use in certain regions, and different regional regulatory landscapes all add extra complexity and scope. At Covance, we are well aware of the global nature of the challenge and are focused on global harmonization. That means using the same robust methods, the same SOPs and quality systems – even the same laboratory information management systems – across the company, which is no mean feat. From a regulatory point of view, even more challenges emerge. We know that there are different maximum residue limits and different compounds in use around the world, but pesticide residue analysis is more than just meeting the appropriate regional regulations. Global companies – and our clients – are increasingly interested in measuring everything, in everything, from everywhere – setting global specifications based on the strictest requirements in each case. Our target lists are growing...
For regulatory and contract labs, strange (and sometimes unknown matrices) are a regular occurrence – especially when it comes to botanicals and other supplements. And though analyzing an unknown sample for (known or unknown) pesticides is clearly an extreme case, it does highlight a challenge that will not go away: the matrix. Perhaps more importantly, it also highlights a trend; gone are the days when cereals, fruits and vegetables were the mainstay of analysis. The matrix challenge appears to be an ever-increasing circle that began with produce, grains and oils, and then expanded to include specialized matrices, such as spices, tea, cocoa, and so on. Today, the circle has grown bigger still, with herbal drug mixtures, dietary supplements... The list continues – as does the complexity.
Maintaining quality in the mayhem
In our labs, we use the SANCO guidelines for pesticides analysis both for validation and routine quality control as a minimum. The importance of quality control, particularly for difficult matrices, cannot be understated. In these difficult matrices, quantitation accuracy can represent a significant challenge, because unknown matrix effects can potentially affect sample preparation (recovery) and quantification (signal suppression/enhancement). Clearly, in all walks of analytical life, identification of contaminants is of paramount importance. Just the presence of certain unexpected contaminants could have huge economic implications (and actually make quantification unnecessary in some cases). Conversely, the quantification of a wrongly-identified compound is entirely pointless. In short, we need very high confidence in our results. For identification with MS/MS, SANCO/12571/2013 states that the minimum should be:- ≥ 2 product ions
- ± 30 percent maximum relative tolerance for ion ratios.
What do targeted and non-targeted really mean?
There appears to be a slight lack of consensus on the meaning of targeted and non-targeted – at least in my experience. From a holistic standpoint, you can consider the difference as two simple questions:- Targeted: is compound X in the sample?
- Non-targeted: what is in the sample?
- Known knowns: targeted processing of targeted – or non-targeted – acquisition data, using analyte-specific conditions (retention time, MRM or selected ions) in the data processing method created with reference standards.
- Known unknowns: (non-)targeted processing of non-targeted acquisition data, using database/library search (fragment match, structure correlation, accurate mass) to get presumptive identification.
- Unknown unknowns: non‑targeted processing of non-targeted acquisition data, using chemometric (differential or statistical) analysis, followed by identification of compounds of interest. A little like trying to find a needle in the haystack.