“For the first time in history, the stability of nature can no longer be taken for granted […] Never has it been more important to understand how the natural world works and how to help it.”
The opening lines of the critically acclaimed Netflix series "Our Planet," narrated by the famous British natural historian Sir David Attenborough, should really make us all stop and think. To play our part, we believe we can contribute to a better understanding of nature with environmental metabolomics.
For the uninitiated, metabolomics is the scientific study of the metabolome – the sum of all small-molecule intermediates and end-products of metabolism found in a given biological sample. Crucially, the composition of the metabolome is not only dependent on the biological system’s current state but is also affected by many exogenous factors, including chemicals – medicines, food components, contaminants, and so on (1, 2).
Intelligent study designs, alongside modern analytical chemistry and sophisticated data analysis, enable us to extract valuable information from the metabolome – a point proven by the application of metabolomics to the area of human health and its significant contribution to the development of personalized medicine (3). It is now beyond any doubt that metabolomics adds an important layer of information to genomic and proteomic analyses.
At the Research Group Metabolomics of the Leiden Centre for Applied Bioscience, our ambition is to apply metabolomics in the fields of ecology, biodiversity, and environmental science. When metabolomics is applied to environmental questions – in other words, used to study the interaction between organisms and external stressors – it is fittingly called environmental metabolomics, or eMetabolomics for short (4). We broaden this definition and include interactions within and between species and their environment, with a view to addressing ecological and biodiversity issues.
The field of eMetabolomics offers promise for the following reasons:
- Changes in the metabolome are likely to be a more sensitive indicator of external stressors than information at the gene and protein levels (4).
- eMetabolomics is sensitive to the environmental effects of (emerging) chemical pollutants, including those not yet routinely monitored and/or with unknown modes of action. eMetabolomics may offer the first mechanistic insights into unknown modes of action.
- eMetabolomics can be used to study specific processes between organisms.
- And eMetabolomics can aid in the rapid identification of organisms.
To stretch the promise of eMetabolomics even further, we focus on the “community metabolome” – the collection of metabolites that are secreted into their environment by all the organisms within a given ecosystem. In essence, the community metabolome reflects the entire influence of various factors (climate, pollution, and invasive species) on the ecosystem in which we live (see Figure 1).
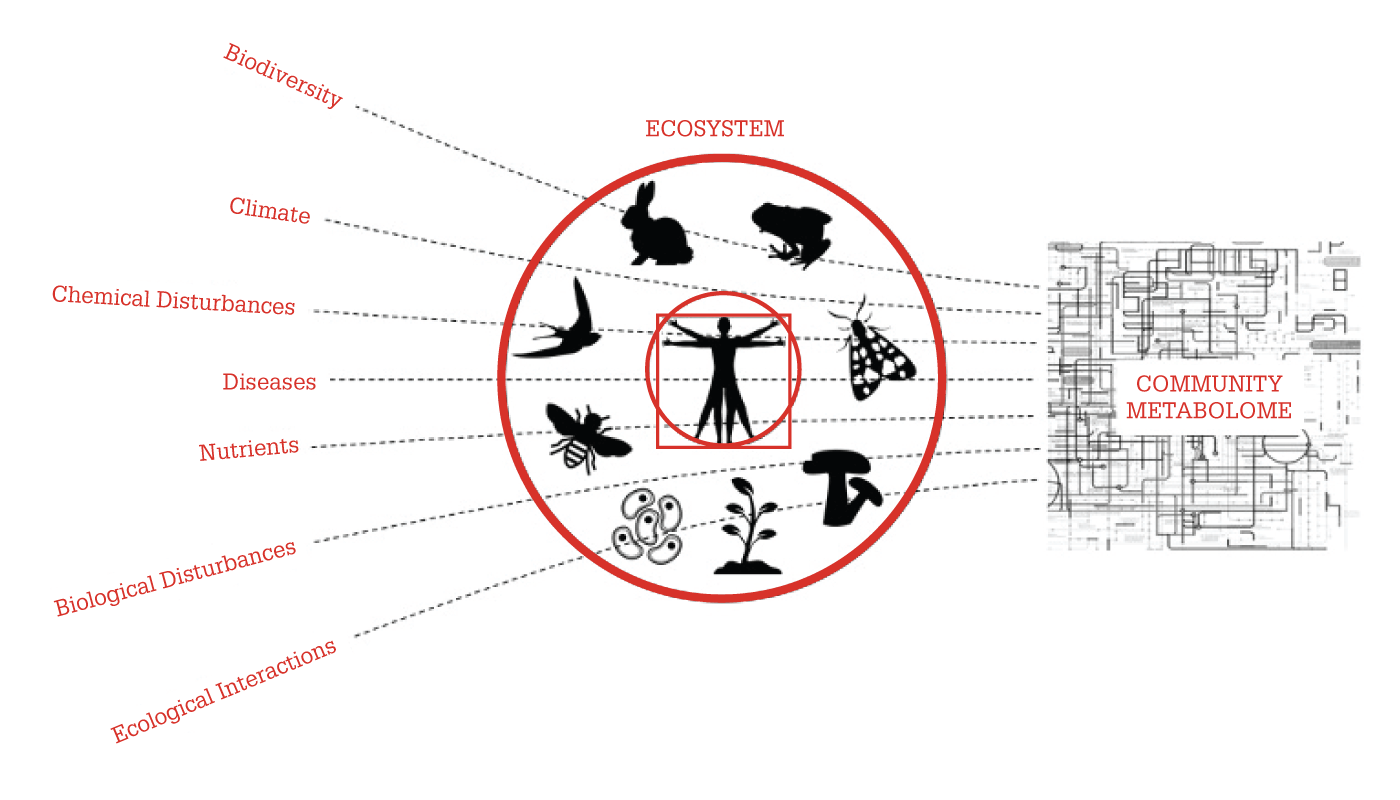
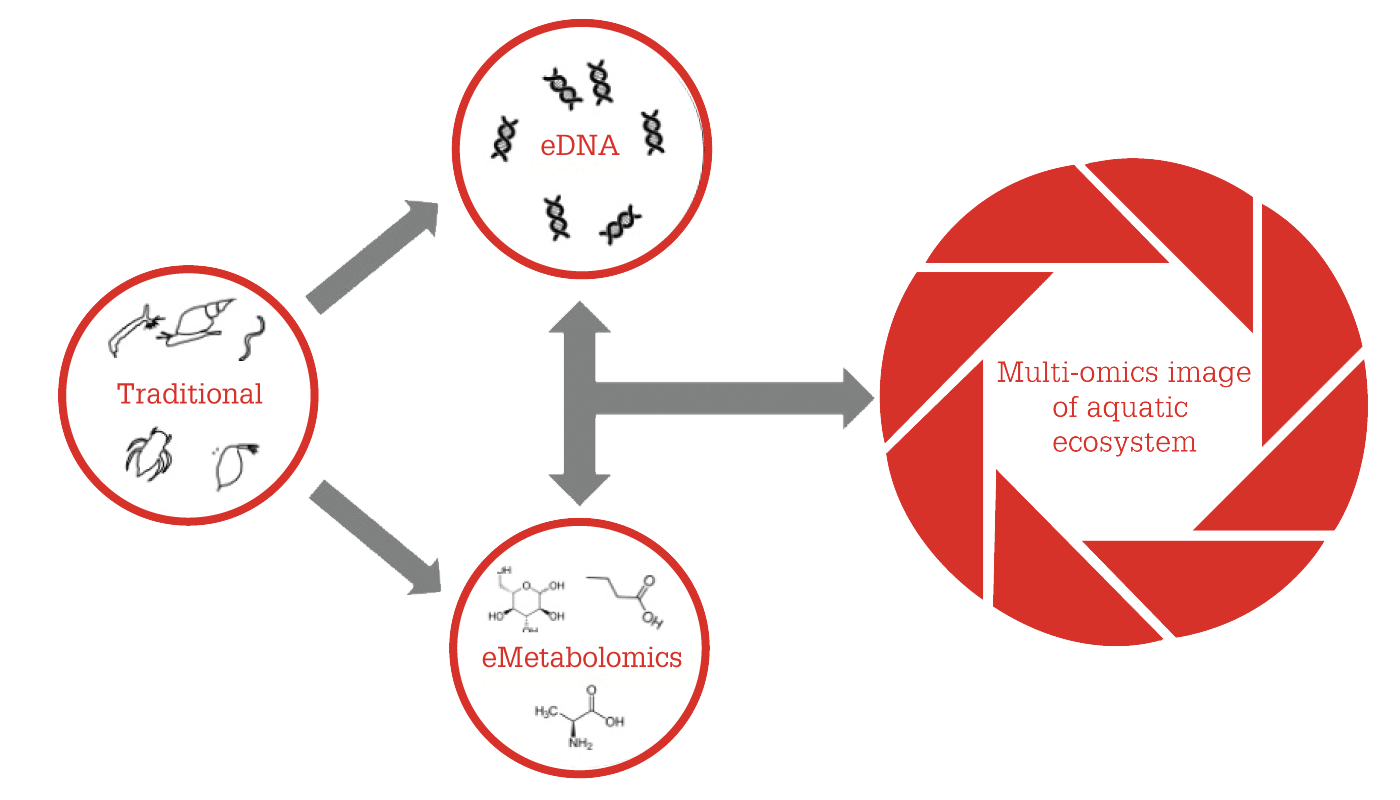
Currently, we are investigating the strength of eMetabolomics at the community level in a multi-omics investigation into ecological water quality. Acknowledging the importance of information layers, we also measure the unique traces of DNA generated by each organism in the aquatic ecosystem – dubbed environmental DNA or eDNA – as part of the project. Combining these two layers of information creates a multi-omics image (see Figure 2). The eDNA analysis provides us with information on which species are present in an aquatic ecosystem, while eMetabolomics provides us with information on which processes are taking place. For example, with the help of eDNA, we can detect the presence of water fleas; with eMetabolomics, we can demonstrate that the water fleas reproduce.
When an ecosystem is affected by a chemical (or any other) stressor, it may deteriorate only slowly – almost imperceptibly – at first. But it is known (5) that the slow and steady decline is very often followed by a period of much more rapid deterioration (like walking down a gentle grassy slope towards a cliff). Once the ecosystem has deteriorated too far – or dropped off the edge of the cliff – it takes a considerable amount of effort to restore its healthy state.
Clearly, a measuring system that can detect disturbances in an aquatic ecosystem at these earlier (and slower) stages of deterioration would be highly valuable – a warning shot. In alignment with metabolomics applied to human health issues, we believe that eMetabolomics is capable of detecting changes in the community metabolome earlier than classical methods used to measure ecological water quality (which is to say, the actual identification and counting of species). To return to the example of the water fleas, prior to dying and disappearing, water fleas first become ill – and that results in a change to their metabolome.
Our current eMetabolomics project aims to understand the effects of both biological and chemical disturbances on an aquatic ecosystem located in the so-called Living Lab of Leiden University. To increase our chances of success, we have built an exciting multidisciplinary research consortium, involving biologists from the Dutch National Biodiversity Center Naturalis and environmental scientists from the Institute for Environmental Sciences of Leiden University. We’ve also recruited the assistance of several ecological consultancy companies and governmental organizations.
The Living Lab consists of a testing field with 36 ditches. Some of these ditches have been disturbed by adding red swamp crayfish (Procambarus clarkii) or the pesticide thiacloprid. The red swamp crayfish is an invasive exotic species that is currently common in many locations, including the western part of The Netherlands, and it is feared to have a negative effect on aquatic biodiversity. Thiacloprid is a neonicotinoid pesticide, which has been shown to have a very negative effect on the insect population and insect-eating birds (6).
Over the course of several months, all of the ditches have been monitored manually, and samples have been collected for eDNA and community-metabolome analyses. First, we will pinpoint metabolites that are linked to disturbances of the aquatic ecosystem in the samples that were collected at the very end of the field study (when the aquatic ecosystems are disturbed maximally). We will then quantify the same metabolites in much older samples, with a view to identifying how early on we are able to detect disturbances. Our expectations? The negative effects of both thiacloprid and red swamp crayfish can be identified early on – during the slow deterioration phase – in the ditch eMetabolome (4, 7).
Our study is certainly specific, but we hope the project will demonstrate the strengths of eMetabolomics more broadly. Equally, we know it will challenge us to seek working solutions to four serious challenges:
- Metabolite identification. The human metabolome database currently contains 114,008 metabolites (2). A community metabolome (the result of many organisms living together in an ecosystem) will likely contain many more metabolites – and a database is currently lacking (7).
- Establishing causal relationships between specific metabolites and specific organisms in a highly complex sample, such as the community metabolome, will not be straightforward.
- Measuring the community metabolome. In doing so, we must develop innovative, on-site sampling techniques and specific preparation methods for various (and sometimes new) sample types (surface water, air, organisms).
- Education. Because (e)Metabolomics is a relatively new but rapidly growing research area, educating and training new technicians and researchers is important. At the University of Applied Sciences Leiden, we have developed a degree course in metabolomics specifically to address this final challenge.
By forging a stronger link between scientists (analytical chemists, environmental scientists, and ecologists) and professional bodies (governments, ecological consultancy firms, water boards), we believe our research and education can help contribute to a healthier living environment – one in which both people and nature can coexist in the best of health.
References
- KL Griffin, “Metabolic profiles to define the genome: can we hear the phenotypes?” Philos Trans R Soc Lond B Biol Sci, 359, 857 (2004). DOI: 10.1098/rstb.2003.1411
- DS Wishart et al., “HMDB 4.0: the human metabolome database for 2018,” Nucleic Acids Res, 46, D608 (2017). DOI: 10.1093/nar/gkx1089
- RD Beger RD et al., “Metabolomics enables precision medicine: ‘A White Paper, Community Perspective,’” Metabolomics, 12, 149 (2016). DOI: 10.1007/s11306-016-1094-6
- BP Lankadurai et al., “Environmental metabolomics: an emerging approach to study organism responses to environmental stressors,” Environ Rev, 21, 180 (2013). DOI: 10.1139/er-2013-0011
- SP Davies & SK Jackson, “The biological condition gradient: a descriptive model for interpreting change in aquatic ecosystems,” Ecol Appl, 16, 1251 (2006). DOI: 10.1890/1051-0761(2006)016[1251:tbcgad]2.0.co;2
- “’Catastrophe’ as France’s bird population collapses due to pesticides,” The Guardian (2018). Available at: https://bit.ly/35WuufX
- MG Miller, “Environmental Metabolomics: A SWOT Analysis (Strengths, Weaknesses, Opportunities, and Threats),” J Prot Res, 6, 540 (2007). DOI: 10.1021/pr060623x




