Two-dimensional techniques are shifting the landscape of analytical science – ramping up levels of data acquisition and offering unparalleled insights into the phenomena we investigate. Our thirst for “more” – especially when analyzing highly complex samples – has driven us out of the first dimension in multiple techniques; 2D NMR spectroscopy has been around for decades, and various flavors of multidimensional chromatography have proven utility in a range of applications. But now, it’s time for the spotlight to shift. Prepare to enter a new dimension – of MS.
Peter O’Connor
“I’m a Fourier-transform ion cyclotron resonance (FT-ICR) mass spectrometrist, and Associate professor at Boston University School of Medicine and Professor of Chemistry at the University of Warwick. Our work has focused on bringing 2DMS to the analytical mainstream, with the hope of developing routine applications in chemistry and biology.”
Dalton Snyder
“I was lucky enough to conduct research under Arlen Kauffman during my undergraduate studies. Arlen introduced me to Graham’s work on ambient ionization and molecular imaging, and I eventually became an “Astonite” myself, developing novel ion trap methods for miniature and portable mass spectrometers.”
Maria van Agthoven
“I’ve worked on several 2DMS projects in labs from Florida State University to the Université de Lille Sciences et Technologies and the University of Warwick. Now, I’m based at the University of Innsbruck, Austria, where I’m continuing this work. The hope? That 2DMS will one day become as commonplace as other analytical techniques.”



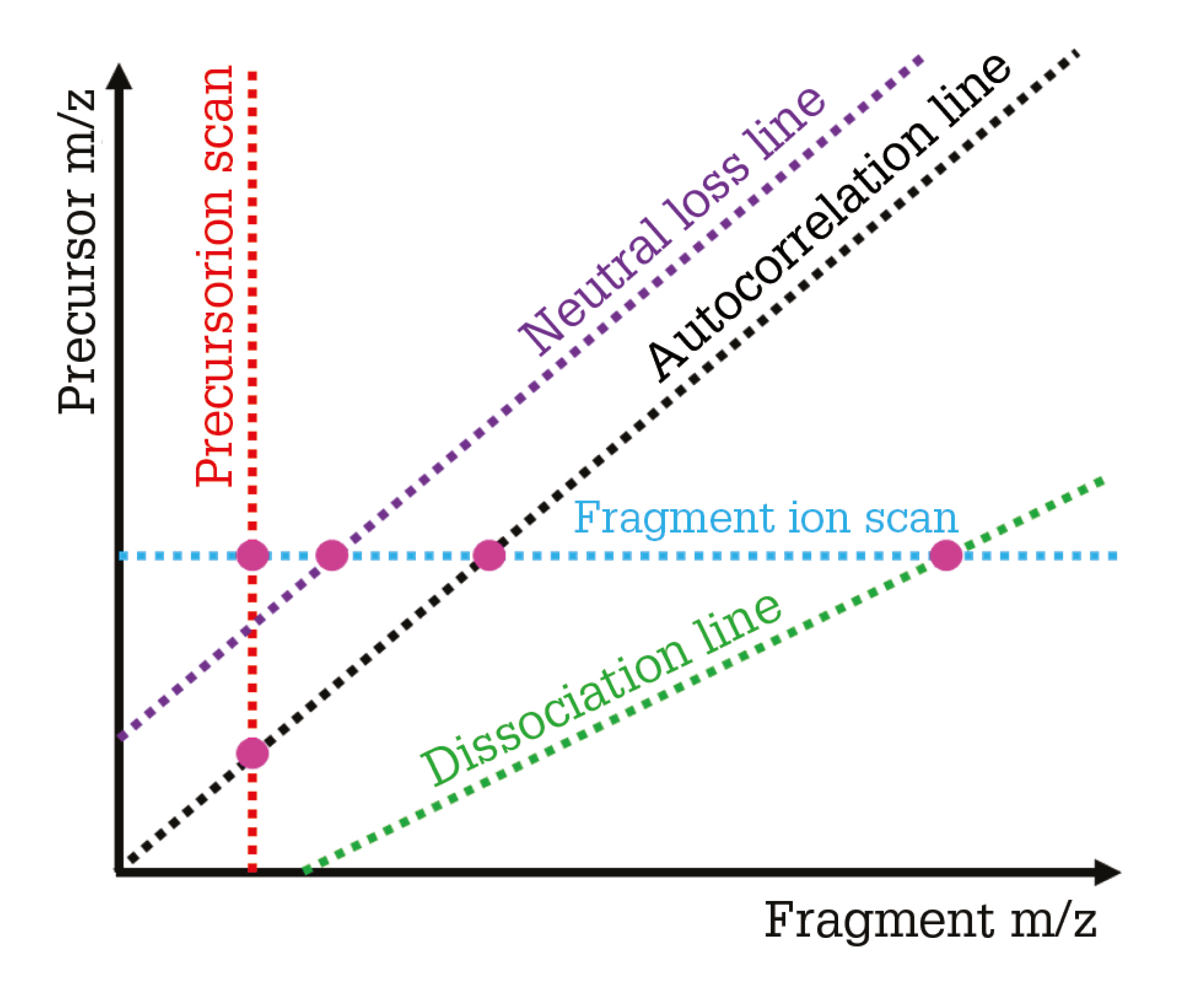
Maria: Ions rotate with a frequency that depends on their mass-to-charge ratio in a magnetic field. This rotation – dubbed “cyclotron motion” – is measured in a high-vacuum, high-magnetic-field cell, such as the ICR cell of an FT-ICR mass spectrometer, and the signal is transformed first to obtain the frequencies, and then the mass-to-charge ratios. In 2DMS, ion radii in the ICR cell are modulated according to their mass-to-charge ratios using a pulse sequence developed by Tino Gäumann and Geoffrey Bodenhausen in the 1980s and inspired by 2D NMR. Because the fragmentation efficiencies of each precursor ion depends on its radius, the abundance of fragment ions also depends on precursor ion radius, which in turn depends on the mass-to-charge ratio; this establishes a correlation between precursor ions and their fragments, which, after a double Fourier transformation, can be mapped on a 2D mass spectrum.
2D mass spectra contain not just the mass spectrum of the analytes from the sample (shown by the autocorrelation line on Figure 1) like a traditional mass spectrum, but also the fragmentation pattern of every precursor ion (the fragment ion scan), the precursor pattern of every fragment ion (the precursor ion scan), and neutral loss and dissociation lines that can be used to find classes of ions in the sample that fragment in identical ways – indicative of similar structures (protein forms with different post-translational modifications, for example).
Peter: In MS, the user first measures the masses of molecules and then performs what is called an MS/MS or Tandem MS study, in which molecules of interest are first isolated, then fragmented (by collisions with neutrals, photons, or electrons), and then a mass spectrum is measured of those fragments. To analyze everything in the sample requires sequentially isolating each precursor molecule, fragmenting it, and measuring the spectrum of the fragments from each precursor.
As Maria says, 2DMS allows us to measure all fragments from all precursors simultaneously. This is done by use of a signal coding trick, whereby the precursors are each modulated in and out of a fragmentation “zone.” Because the fragments are only created inside that “zone,” the fragments also modulate in their intensities according to the modulation frequency of the precursor. Thus, we know exactly which fragment is derived from which precursor in a complex mixture. 2DMS is therefore particularly useful for complex mixtures, or mixtures that are difficult to separate. Good examples include polymeric samples, proteomics samples and very complex mixtures like lignin, biofuels, and petroleum – we are currently focusing on such analyses in my lab.
Peter: 2DMS is actually an “old” technique in that it was initially demonstrated in 1987 (1), but was subsequently ignored for many years due to computational limitations – we simply couldn’t process the volume of data produced. In 1990, a 2-megabyte Fourier transform would take about 30 minutes, but around 1000-4000 of these calculations are needed to produce a 2D mass spectrum. By 1998, the CPU of a standard desktop PC could perform these calculations in half a second – though it would take a lot longer to transfer the data from hard disks into RAM. Continuous improvements in computational capability from then until now (Moore’s Law estimates that computing power doubles every two years) mean that we can use cluster computers to parallelize the Fourier transform and data analysis, or simply use a maxed-out desktop PC with 256 gigabytes of RAM and multiple cores for data processing. 2DMS is now a credible approach for conducting analyses in a standard MS lab.
We have also developed a new way to conduct 2DMS analyzes on a linear ion trap, making the approach amenable to standard quadrupole time-of-flight mass spectrometers. Maria, Christopher Wootton and I then commercialized this success by setting up a startup company called Verdel Instruments with the University of Warwick.
Dalton: We weren’t the first group to use 2DMS, but it had previously been compatible only with FT-ICR platforms. Peter (as you can probably tell from the above!) has been a key driver of the technique, having conducted pioneering research for several years. His work developing a method for 2DMS analysis on FT-ICR instruments laid the foundation for my research in the area, which represented a significant change in thinking about how 2DMS could be conducted. 2DMS on the FT-ICR consists of a series of scans from which the tandem MS data domain can be reconstructed (which takes hours), while the technology developed by Graham, Lucas and myself uses one scan, taking around one second to complete. Of course, the resolution of the FT-ICR is significantly higher, but we trade resolution for instrument runtime.
Maria: I learned about FT-ICR MS during my first postdoc at Florida State University, and was hired by Christian Rolando to enable 2DMS on a Bruker instrument in 2009. We showed that 2DMS was viable with laser-based and electron-based fragmentation methods and we optimized the pulse sequence parameters for maximal signal-to-noise ratio, after which I moved to the University of Warwick to work alongside Peter O’Connor. At my current institute – the University of Innsbruck – I work on top-down RNA and histone analysis. We study these molecules because they are often subject to post-translational molecular modification, and thus are difficult to separate with chromatographic approaches.
In this space, I particularly focus on the development of quantification techniques in 2DMS, based on the neutral loss lines and dissociation lines demonstrated. Data analysis techniques will be invaluable in 2DMS research due to the sheer volume of data produced. Some of the analytical information present in 2D mass spectra that cannot be obtained with any other method. Precursor and fragment ions are correlated not only through precursor mass-to-charge ratios, but also through precursor charge state. Information on fragmentation mechanisms can also be uncovered by analyzing the harmonics present in the spectra.
Dalton: The initial applications that we looked at for 2DMS were relevant to our collaborators at NASA and FLIR. With FLIR, we focused on the detection of opioids (largely fentanyls) and other nefarious substances to demonstrate forensic utility. With NASA, we used 2DMS for the detection of amino acids and other small organics – molecules that we hope to find on Mars (or elsewhere) one day. Regarding the latter application, the hope is that NASA will be able to send a 2DMS-equipped ion trap to Mars or extraterrestrial icy moons to deduce the organic chemical makeup of a rock or ice sample, from which they could infer the past existence of or present suitability for life.
“Many pressing issues in science and technology have at their heart the question of molecular composition. Most real samples are complex mixtures and most real problems call for quick decisions based on molecular information. This statement is as true of the trace residues on the fingers of a passenger boarding an aircraft as it is of the spatial distribution of complex phospholipids in the brain tissue of a glioma patient as it is of the bacterial composition of lettuce in the quick-pick line. An argument can be made that the greatest need in technology is for near instantaneous chemical identification tools. Ion traps fitted with ambient ionization sources and performing 2D MS/MS scans come close to answering this challenge. The near future for 2D MS/MS must be focused on utilization of the existing capabilities in solving real problems.”
Graham Cooks, Henry Bohn Hass Distinguished Professor of Chemistry in the Aston Laboratory of Mass Spectrometry, Purdue University
The limitations imposed by miniature mass spectrometers in terms of size, power, weight, and sample consumption were the main factors underscoring our research. Initially, we developed precursor and neutral loss scans (2) (3) as efficient means of sorting through complex chemical mixtures using a simple yet sensitive portable ion trap mass spectrometer. These methods were thought either impossible or problematic on anything but a bulky, power-hungry multiple analyzer instrument like the triple quad. 2DMS is an extension of our prior work, wherein all possible precursor scans, neutral loss scans, and product ion scans are conducted in a single run. This approach allows us to probe mixtures and obtain structural information on virtually every compound in the sample quickly (minimizing power consumption) and with minimal sample consumption.
Peter: 2DMS is still in its infancy with respect to technical applications, but there are some clear areas where it will have an advantage over alternative methods. Proteomics is an interesting case that my team has been exploring in detail – inspired by the potential for 2DMS to explore complex protein mixtures, as showcased by the “Uncoiling Collagen” paper from Simon and colleagues in 2016 (4). The team used 2DMS to obtain data on trypsin digests of collagen cleanly and clearly using FT-ICR MS and a blind, unoptimized method. We have continued this work by proceeding onto more complex bulk proteomics experiments. On this front, we’ve obtained many interesting, unpublished results that are making their way out of the lab… But you’ll have to wait until those are published! As a sneak preview, they involve 2DMS for whole protein analysis, polymers, pesticides, and – of course – proteomics.
Dalton: The technology is still fairly niche at the moment. Our technology, for example, has only been developed in the last couple of years, so it has much room to grow and evolve – hopefully leading to improved performance metrics, such as mass spectral resolution, sensitivity, and speed. I would say the biggest developments in the field to date are the initial conception of 2DMS on the FT-ICR by Pfändler and colleagues (1), optimization of the pulse sequence and data analysis technologies by the Rolando group, and our initial conception of 2DMS on quadrupole ion traps. At the moment, 2DMS on quadrupole ion traps is capable of near-unit precursor ion resolution (up to ~120 product ion resolution, variable with mass-to-charge) and a limit of detection that compares quite favorably with ion trap full scan mode.
Maria: Today, 2DMS can be used for data-independent analysis in research laboratories for tandem mass spectrometry experiments without ion isolation. The pulse sequence can be optimized on any FT-ICR mass spectrometer, denoising algorithms enable easy data analysis and interpretation, and Marc-André Delsuc and his group have even developed an open-source program package for data processing, visualization, and analysis. So far, 2DMS has been used for bottom-up and top-down proteomics, polymer analysis, and small molecule analysis. But I believe the best is yet to come.

Maria: I think we’ll see increased use of 2DMS in the FT-ICR MS community in the next 5–10 years, particularly for quantification studies in top-down and bottom-up proteomics. The methods developed for 2D FT-ICR MS can then be transferred to 2DMS on a linear ion trap with time-of-flight mass analyzers as they reach the market. These instruments will be cheaper and faster (albeit with lower performance in terms of resolution and mass accuracy), and will be potentially coupled with chromatographic techniques. Once we can use 2DMS to separate analytes by chromatographic time, mass-to-charge ratio, and charge state in a fully multiplexed way that does not involve loss of sample through isolation, we will have overcome a major analytical barrier. In short, I anticipate that 2DMS will allow us to extract more information from highly complex samples – perhaps even by several orders of magnitude – in the years to come.
Dalton: The truth is that we have little idea what the future holds for 2DMS. And that’s the exciting thing about this research! In the next several years, I imagine we will see new versions of the technology emerging with improved resolution, sensitivity, and scan speed. Perhaps someone will think of clever ways to do 2DMS on quadrupoles or time-of-flight instruments, too. I expect further interest from those involved in the development of new portable and miniature mass spectrometers and perhaps 2DMS will end up as a key feature of the next generation of devices!
I also hope to see more researchers getting involved in instrument development for 2DMS. So far, the community has only covered two types of spectrometer (quadrupole ion traps and FT-ICRs), so there are many more to go. I certainly imagine new methods of 2DMS emerging as more clever graduate students get involved in the subject.
Peter: The technical limitations of 2DMS are mostly well understood, but there’s a persistent problem in calibration of the vertical axis that remains. This problem is related to the timing of the detection electronics and is annoying to define. For this reason, every publication to date has had to use internal calibration of the vertical axis, but we believe that some relatively simple modification to the electronics will eliminate this calibration problem and allow robust, external calibration of the vertical axis of 2DMS data.
Beyond this calibration problem, the main technical problem remains processing the data obtained. We are developing the software to do the 2D Fourier Transform (routine), the denoising (tricky and under development), peak picking (challenging, but conceptually straightforward), and the assignment of masses from the resulting peak lists to important chemical features (an ongoing and probably perpetual struggle). Still, if we can push most of the technical problems towards being software problems, many more individuals can become engaged in the solution – and that can only be a good thing for the field.
References
- Pfändler et al., “Two-dimensional fourier transform ion cyclotron resonance mass spectrometry”, Chem Phys Lett, 138, 195 (1987). DOI: 10.1016/0009-2614(87)80367-6
- Snyder DT et al., “Single analyzer precursor ion scans in a linear quadrupole ion trap using orthogonal double resonance excitation”, J Am Soc Mass Spectrom, 28, 1929 (2017) DOI: 10.1007/s13361-017-1707-y
- Snyder DT et al., “Implementation of precursor and neutral loss scans on a miniature ion trap mass spectrometer and performance comparison to a benchtop linear ion trap”, J Am Soc Mass Spectrom, 29, 1355 (2018). DOI: 10.1007/s13361-018-1922-1
- Simon HJ et al., “Uncoiling collagen: a multidimensional mass spectrometry study”, Analyst, 141, 157 (2016). DOI: 10.1039/c5an01757b




