Pieter Dorrestein’s research aims to understand at the chemical level how microbes interact with each other, with their surroundings, and with their host. “We are interested in the specialized molecules that microbes produce to curate their specific environmental niche,” he explains, with the goal of “repurposing them for clinical applications,” citing the examples of antibiotics (Penicillin, erythromycin, daptomycin), immunosuppressants (rapamycin), and anti-cancer agents (epothilones).
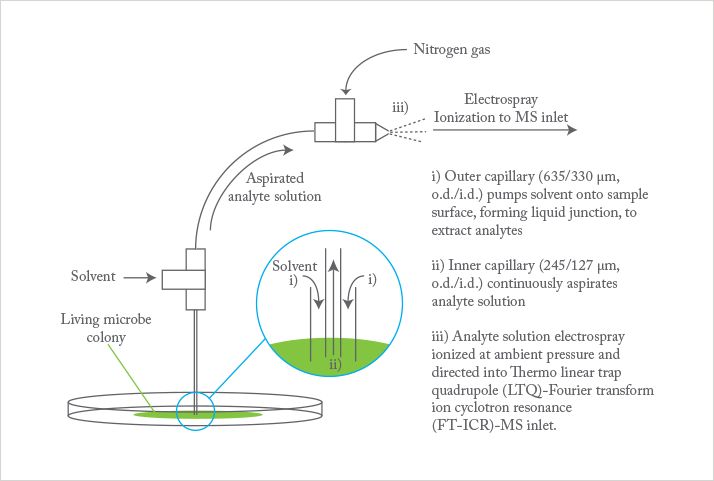
To achieve their goal, researchers in Dorrestein’s lab at the University of California, San Diego are directly sampling analytes of living microbe colonies ahead of electrospray ionization (ESI) and tandem mass spectral analysis (1). The key component is a liquid microjunction surface-sampling probe (LMJ-SSP). It is based on a device developed by Van Berkel at Oak Ridge National Laboratory, which comprises two co-axial capillary tubes that simultaneously deliver and aspirate solvent (see Figure 1).
“Current abilities in sequencing allow us to discover the genetic capacity of microbes to make such specialized metabolites, but the analytics to detect them is far behind. Traditionally, it is done one molecule at a time.” By removing the need for sample preparation and studying living colonies, the new technique could open doors to high-throughput studies or be used to probe novel microbiome interactions. When asked about sensitivity, Dorrestein replies: “The method is not as sensitive as LC-MS because we only detect the most ionizable or abundant molecules. But it turns out that many of the specialized molecules that alter the environmental niche of microbes are produced in fairly large quantities”. In fact, it is not uncommon that the compounds of interest are produced in tens or even hundreds of micrograms per ml in a Petri dish, ranking them among the most concentrated molecules in the sample. “The localized nature of the sampling method further enhances observed concentrations because the molecules are often highly localized as well,” he says.
Given the diverse nature of the chemical landscape uncovered by the approach and the complexity of the datasets produced, intelligent data analysis is essential. By considering compounds and their spectra in the context of a network of chemically similar molecules, “molecular families” emerge, which are represented by a tight cluster within the molecular network. Such molecular networking can be used to form a system-wide picture or to characterize individual compounds. Furthermore, combining molecular family information with gene cluster data generates tools, such as peptidogenomics, that makes high-throughput analysis a real possibility. An earlier version of the technology – nanoDESI – combined with molecular networking (3) piqued the interest of Roberto Kolter and Matt Traxler back in 2012 (4). They lined up potential question-based applications, including “What chemical ecology underlies the microbiome associated with human skin? What signals potentiate relationships between hosts and symbionts? How do soil bacteria, long known to make a variety of antibiotics, deploy their arsenals in the presence of competitors?” The Dorrestein approach has some bugs: capillary clogging needs to be reduced and depth recognition for automation must be added. When these issues are addressed (solutions are in the pipeline), progress on answers to Kolter and Traxler’s questions, and more besides, will become a real possibility.
For more on research at the Dorrestein Lab’s, visit: chem-faculty.ucsd.edu/dorrestein
References
- C. Hsu et al., “Real-Time Metabolomics on Living Microorganisms Using Ambient Electrospray Ionization Flow-Probe”, Anal. Chem. 85, 7014−7018 (2013) doi: dx.doi.org/10.1021/ac401613x http://web.ornl.gov/sci/csd/Research_areas/obms_group.html J. Watrous et al., “Mass spectral molecular networking of living microbial colonies”, PNAS 109 (26), E1743-E1752 (2012). M. Traxler and R. Kolter, “A massively spectacular view of the chemical lives of microbes”, PNAS, 109 (26), 10128-10129 (2012).
