How did you come to work with microchip systems?
Sabeth Verpoorte: I started out in 1990 as a postdoc at Ciba Geigy AG (in the lab of Andreas Manz, one of the pioneers in the field of microchip systems). There, we demonstrated various chip-based applications using devices fabricated elsewhere. We were end-users of fabrication technologies, and provided microfluidic technology to the company.
In 1996, I joined a lab headed by Nico de Rooij, with the goal of realizing various analytical principles in the miniaturized chip format. We and others produced chip-based devices that incorporated multiple sample processing functions into a single device, sometimes in a highly parallel fashion. The unique flow properties that exist in small channels allows functionality that simply isn’t possible in larger-volume flow systems. In 2003, I assumed a chair in a pharmacy department at the University of Groninge, taking me far outside of my analytical and technological “comfort zone” – but I felt I would only truly understand how microfluidics can solve problems in the life sciences if I placed myself in that environment.
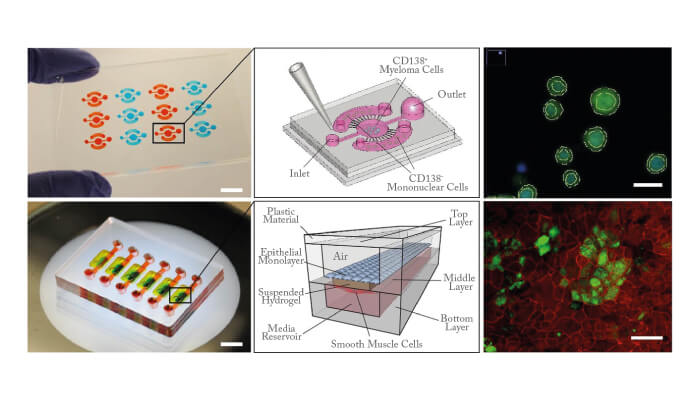
Edmond Young: I started working in microfluidics when I began my PhD studies at the University of Toronto in 2004. At first, I studied electrokinetics in microchannels, but in 2005 I began studying how to culture endothelial cells in microfluidic systems for cardiovascular research, which became the focus of my doctoral thesis. In 2008, I moved to the University of Wisconsin-Madison to do postdoctoral research with David Beebe, where I became interested in applying microfluidic systems to cancer biology. During my postdoc, I designed the MicroC3 platform, a microfluidic chemosensitivity and resistance assay for testing drug responses of multiple myeloma tumor cells (1) (2). This technology relies on microscale geometric features to coculture tumor cells and non-tumor cells from the same patient. It is currently being used by a startup company LynxBiosciences, Inc. for rapid analysis of treatment response in blood cancers.

Jaap den Toonder: I started working with microchip systems in 2004 as a principal scientist at Philips Research laboratories in the Netherlands – initially i n t e g r a t i n g PCR and immunoassays in microfluidic format for nucleic acid and protein diagnostics. This work resulted in a nucleic acid testing system (Idylla, commercialized by Biocartis; www. biocartis.com. I continued to work on microfluidic systems, first as a part-time professor (next to my Philips job) and from 2013 as full professor at Eindhoven University of Technology. By 2010, I had become very interested in “organ-on-a-chip” technology, and co-organized an international workshop in Leiden on this topic. This led to the founding of the Dutch organ-on-a-chip institute (hDMT; www.hdmt.technology). Here, research groups from technological, biological, and medical backgrounds team up with biotech companies to develop organ-on-a-chip systems.
What are you working on right now?
Toonder: My focus is on developing novel technical solutions to create microfluidic control, as well as representative environments for advanced cell culture. In collaboration with our partners, we apply these advances to diagnostic and organ-on-chip applications; for example, “artificial cilia” – magnetic microactuators that can be integrated in microfluidic chips to pump and mix fluids, as well as manipulate particles or cells (1). We also use magnetic particles to induce mixing or function as microcarriers for molecules (2), and develop microfluidic systems to analyze the mechanical properties of single cells, which can be a relevant marker for various diseases (3). Finally, the fastest-growing research topic in my group is “organ-on-a-chip”. Our main focus is on developing microfluidic chips that allow us to study the main steps in the process of cancer metastasis. We are most interested in the effect of microenvironmental factors on the invasion and migration of cancer cells in tissue (4).
Young: We are developing a lung-airwayon- a-chip (3). This is fabricated entirely from poly(methylmethacrylate) (PMMA or acrylic), has an arrayable format, and uses a unique “floating” hydrogel within the device to create upper and lower cell culture compartments. The floating hydrogel also serves as the basement membrane between the airway epithelial cells and bronchial smooth muscle cells cultured on opposing sides of the gel. So far, we have demonstrated that the airway-chip can remain viable for over a month, and that the airway epithelium can produce mucosal proteins similar to airways in vivo. We are hoping to use it in air pollution studies in the near future.
Honing: My research is mainly focused on the hyphenation and integration of analytical detection technologies in microfluidic systems. Developing miniaturized separation technologies for the online monitoring of (bio)chemical processes in microflow chemical reactors or organ-on-a-chip systems is a fascinating challenge. My personal interest is in designing novel IMS-MS/MS technologies for “universal” detection of all small molecules, together with the engineering and application of novel chemical flow-reactor or organ-on-a-chip devices as part of interdisciplinary research teams.
My second research field relates to the understanding of material–biology interactions; for example, using surface plasmon resonance to measure the release of molecular markers for cell systems being exposed to the release of unknown chemicals from biomedical materials. Here, I collaborate with industry and research groups working on novel biomedical materials and advanced drug delivery technologies.
Why is modeling so important?
Verpoorte: In recent years, there have been significant advances in imaging technology, allowing some biological processes to be monitored inside living human or animal bodies. However, teasing out what is happening on a molecular level remains difficult, as the molecules involved are often “invisible” to imaging systems. The modeling of biological systems ex vivo thus remains our only option for studying many processes. It is vital that the cells or tissue within the model are maintained under conditions that resemble the in vivo situation as closely as possible. By ensuring that the microenvironment is controlled and well defined, it is easier to study a single cellular mechanism against a noisy background of biochemical activity, and thus improve the reproducibility of the biological process in question.
Toonder: To understand the behavior of complex systems as a whole, you need to study the sub-parts in a highly controlled manner. Biological model systems like organ-on-a-chip allow us to take such a “reductionist” approach. All models have limitations – but these do not make them less important or relevant.
Honing: Molecular modeling to calculate “collision cross sections” of molecules in the gas-phase helps us to understand separation mechanisms in ion mobility spectrometry (IMS). The same holds for the calculation of ionization potential, or even proton transfer reaction in ion sources. Crucially, it also allows the modeling of adsorption mechanisms and kinetics in material–biology interactions.
Young: Coming from a mechanical engineering background with experience in theoretical and computational modeling in fluid mechanics, my appreciation is from an entirely different perspective to most in the field. But I still believe a general rule applies to all modeling: a model should be made as simple as possible, but no simpler. I view the modeling of biological systems in the same way, and to address many of the outstanding questions in modern biology, petri dishes and well plates are too simplistic. With the latest technological advances in microfabrication, biofabrication, 3D printing, and computing, the modeling of biological systems should be – and must be – improved so that cell culture models can keep pace.
What are the key applications for lab-on-a-chip systems?
Honing: Pharmaceutical R&D (medicinal and process chemistry, pharmacology and toxicology), food production (dairy production, safety), biomaterials and devices (anti-fouling coatings in catheters, novel dental curing)and many others.
Toonder: As models of human disease for biomarker/drug discovery and development, organ-on-a-chip models provide a more representative context for cell/tissue culture than current in-vitro models, so that organ functions are recapitulated more closely. By using human cells, they likely are also more representative than animal models, and they do not have the ethical issues associated with animal testing. In personalized diagnostics, we see organ-on-a-chip models in which potential treatments can be tested on patient-specific material to help tailor therapies to the individual.
Young: Some other possible applications include testing of environmental pollutants, toxicological testing of chemicals, and – further into the future – nutrition and aging.
What most excites you about this area?
Verpoorte: The field is enormously diverse, and our increasing understanding of how fluids behave in microchannels is enabling new principles that are in turn leading to new applications. Seeing microfluidics being used to develop diagnostic devices for patients in regions where lab facilities are not available is very gratifying. Personally, I derive the greatest pleasure from the exquisite control we can achieve over the movement of fluids, particles and chemical species in confined microspaces. I am continually amazed at the wonderfully original ways in which we can manipulate molecules, particles and cells at scales far too small to see with the naked eye.
Young: We’re at a point in the field right now where there is a lot of excitement – and rightfully so. The feasibility of accurately recapitulating in vivo physiological functions has already been demonstrated for a number of organs, and the evidence continues to grow. Much research still needs to be done, and we should be cautious not to overhype the potential of the field, but I think there is a great opportunity here to develop and establish a new class of in vitro models that will be far more representative of real-life tissues than conventional in vitro platforms. What excites me the most is the idea of seeing organon- a-chip devices being used in the future as commonly as we see well plates being used today.
Toonder: I’m enthusiastic about the merging of technology and biology. In organ-on-a-chip devices, this merging happens literally: the technology provides the environmental conditions needed for the biological processes to happen, which enables us to answer biological questions that cannot be addressed otherwise. Working on technology that gives insight into the causes of disease and may lead to novel breakthrough solutions for treatments is satisfying; on the other hand, biology is also an inspiration to develop novel technology (for example, bioinspired microfluidic control).
What role does analytical science play?
Toonder: Analytical science is crucial. An organ-on-a-chip model needs to be “interrogated” to understand its behavior. Analytical science provides quantitative (bio)chemical information that is essential for interpreting the response of the model to various cues. In many organ-on-a-chip studies, the information extracted from the models remains qualitative (for example, studying morphological changes). In my view, the quantitative results that can be obtained by the analytical sciences are instrumental in predicting the responses of the model (which would be needed for personalized diagnostic tools, for example).
Verpoorte: Analytical science itself is driven by an exponentially growing demand for information about increasingly complex systems. Without analytical scientists pioneering the use of chips, developments in the microfluidics field may have been much slower. As analytical chemists, we are trained to be exacting. As with any relatively new technology, the measurement procedures implemented with microfluidics must satisfy rigorous criteria with respect to reproducibility and ease-of-use.
Young: I agree that it plays a critical role. Our ability to understand cellular behavior and function relies heavily on our ability to detect, identify, and measure molecules and proteins within and around cultured cells, as well as detect and measure physical, electrochemical, or other signals from cells. A major research theme in the labon- a-chip field has always been the direct integration of analytical instrumentation on-chip, and I think the ability to perform analysis on-chip will be what distinguishes systems with realtime capabilities.
Can an organ-on-a-chip be an analytical system in its own right?
Toonder: If it is appropriately designed and set up, it can be. Although often more complex than classical analytical systems, organ-on-a-chip models can produce a quantitative, multifactorial bio/physico/chemical analysis of the organ model at hand – information that can be used in medical/pharmaceutical practice (treatment strategy choice, drug development and so on).
Verpoorte: In vitro studies invariably involve assessing a cellular readout in response to a medicine, nutrient or toxicant. Cells themselves are the objects of study. When instead the fate of compounds processed by cells is of primary interest, analysis of these compounds and the products produced becomes the experimental focus. The cell cultures in this situation become a n integ ra l pa r t of the analytical process, rather than the object to be analyzed.
Young: An organ-on-a-chip system can include analytical elements, but does not necessarily have to. Any microengineered model system that recapitulates the core functions of a particular tissue or organ may be considered an organ-on-a-chip, with or without analytical instrumentation. How important is multidisciplinary collaboration in this field?
Verpoorte: Microfluidics is a wonderful example of a “cross-over” technology, which very quickly expanded from being analytical chemistry-centric to a much broader applied technology in chemistry, biology and medicine. The fact that the original term “miniaturized total analysis systems (μTAS)” was quickly replaced by more general terms, such as microfluidics and lab-on-a-chip, speaks to the adoption of the technology by researchers in other fields. The crossover success continues with the latest developments in the organ-on-a-chip field, where microfluidics enables – again – the controlled transport of fluids, molecules and cells for the precise microengineering of cellular environments.
Young: In an interdisciplinary field such as this, it is essential to collaborate and bring different ideas together. In the case of the MicroC3 platform, it was developed in collaboration with both a basic biologist and an oncologist during my postdoctora l work at the University of Wisconsin-Madison. In the case of the lung-airwayon- a-chip, we worked with a lung transplant surgeon and an atmospheric scientist (both from University of Toronto). We have partnered with various biotech companies (for example, Bio- Rad, LynxBiosciences) to translate and commercialize the technology or methods we develop. And more recently, I have started collaborating with another oncologist and an immunologist to discuss how organ-on-a-chip technology could be used to study pancreatic cancer.

How will the field evolve in the next 5-10 years?
Toonder: Most current organ-on-chip models are very specific devices operated by experts in specialized labs. But that does not mean we need to make the models simpler; it means that we need to design the technology to be more accessible (a smartphone is an incredibly complex device, but it can still be operated by almost everyone). We need a continuous dialog between the various disciplines involved, and between technology developers and end users.
Verpoorte: There are parallel advances being made in the use of adult stem cells to generate tissue, and in biomaterials that mimic extracellular matrix. I also foresee the incorporation of more analytical functionality into organ-chips. By monitoring important parameters for incubation and cell response, better operational control over organ-chip performance will become possible.
Young: I think a few important changes are on the horizon, based on current trends. Laboratory microfabrication techniques are becoming increasingly accessible, and mass manufacturing of devices is becoming more prevalent. More and more companies are offering microfabrication services, and competition will likely drive down the cost of chip design and manufacturing, making chips more accessible for the non-expert.
Honing: With the development of novel micro-flow reactors, and the rather straightforward upscaling, lab-on-a-chip technology has received major interest from the pharmaceutical industry. Continuous manufacturing regulations (a draft of which has been issued for consultation by the FDA), will open many new possibilities for the design of analytical technologies in miniaturized chemical processing. The paramount role of organ-on-achip technology in understanding cellular signaling, translate cellular response to organoids and hopefully organs, is under discussion.
Young: Organ-on-a-chip systems will need to be rigorously validated with animals before we continue with any serious applications in humans. For example, murine organ-on-a-chip devices for lung, gut, and liver could be directly validated with tissues and organs harvested from lab mice, enabling designs of organ-on-a-chip devices to be further tested and refined. We will likely see increased interest in machine-learning approaches for analysis of different features acquired from organ-on-a-chip-derived data (15). However, I am a firm believer in “garbage in, garbage out,” so regardless of how large our datasets become, they will be of little use if the organ-on-a-chip devices themselves are not accurate representations of tissues in vivo.
Verpoorte: Another fascinating consequence of organchip development will be the realization of multi-organ or microphysiological systems. These systems could be implemented much earlier into the drug development process to identify possible organ interactions leading to toxic drug effects, for instance. This kind of information would allow “go–no go” decisions about the further development of new chemical entities to be made much earlier, saving millions of dollars through circumvention of expensive preclinical and clinical studies – an actual paradigm shift in drug development!
How can analytical scientists feed into the evolution?
Young: Analytical scientists have a major role to play in the continued advancement of this field. One application where analytical scientists will likely have an indispensable role is in metabolomic profiling from organ-on-a-chip systems. Because of the need to handle very small volumes of sample from these devices while detecting increasingly subtle differences between samples, analytical scientists will be tasked to come up with innovative solutions on how to improve instrumentation and processes to more accurately measure an increasing number of metabolites with less and less sample material. As an engineer interested in developing the next-generation of organ-on-a-chip devices with embedded analytical elements, it is critical that engineers and analytical scientists work side by side to achieve seamless integration. I hope analytical scientists continue to stay open-minded and help us realize the full potential of these biomicrofluidic systems.
Honing: To be able to come up with new and novel technologies and methodologies, analytical scientists will need to better understand the needs of their colleagues. With the increasing interest in microfluidics technologies, miniaturization of analytical technologies is needed, without loss of instrumental performance (e.g. resolution) in dynamic systems. We need “process analytical technologies” for complex time-dependent monitoring of biological and chemical processes. For example, 3D printing of drug delivery devices, or even pills, will go hand in hand with the application of new “handheld” miniaturized devices. Although many systems exist, they lack universal detection, are still too expensive, and too new to be integrated with novel data management tools. More specifically, despite the successful application of IR/NIR, fluorescence and UV monitoring, the positive development of benchtop on-line NMR (80 Mhz) and the hyphenation with ESI-MS – the “holy grail” in sensitive, universal detection – is still missing. The latter technology outperforms other technologies in terms of sensitivity, dynamic range or scanning speed. A solution for universal (all molecules, including those who lack “chromophores”), sensitivity (pg/ml), large dynamic range (4 orders of magnitude to accommodate the detection of low abundant impurities) is still missing.
Verpoorte: To help advance chip-based analysis, analytical scientists in this area should continue to do what they do best: making measurements. Continued collaboration with researchers in diverse fields is key to moving microfluidics forward both fundamentally and in practice. For us, interdisciplinary thinking comes naturally. We are thus welltrained to bridge any “scientific cultural differences.” I believe we will continue to play a key leadership role as microfluidics moves into the future.
Sabeth Verpoorte traces the development of microfluidic technology, from simple systems to today's sophisticated organ analogs.
One of the perennial challenges facing analytical scientists is the rapid and efficient generation of highquality information, while rigorously maintaining good analytical practice. That demand for information has been growing over the past decades, and continues to grow at an incredible rate, along with, or perhaps because of, humanity’s improving ability to handle and process information. It is estimated that while the doubling time of medical knowledge in 1950 was 50 years, it had dropped to 7 years in 1980, and to just 3.5 years in 2010. In 2020, it is predicted to be 0.2 years, or just 73 days (8).
Much of the information in the physical, life and medical sciences is generated through measurement. As an old Dutch adage says, “meten is weten” – to measure is to know. And the more we know as scientists, the more we need to measure. It is no surprise, really, that two of the major trends in analytical chemistry over the past few decades have been miniaturization and automation.
Miniaturization of chemical analysis has been driven by the realization that, in most cases the smaller the sample, the easier (and faster) it is to work out the qualitative and quantitative composition of that sample. However, when considering liquid samples, reduced sample volumes often require us to work with sub-microliter amounts, a volume regime which is no longer accessible with laboratory glassware or even micropipettes. In analytical separation techniques, this means working with column or particle diameters smaller than 10 micrometers. Analytical scientists have thus had to resort to other means to work controllably and reproducibly in such small volumes. Enter the microfluidic chip, or lab-ona- chip as it has also come to be known.
Devices are fabricated in planar glass, silicon or polymer substrates using microtechnologies based on those originally developed for the microelectronics industry. Alternatively, polymer fabrication techniques like micromilling and micro injection molding are becoming an increasingly popular route to the fabrication of analytical microdevices. Working with planar substrates enables the formation of complex interconnected microchannel networks containing precisely defined, non-uniform cross-sections and layouts tailored to just about any application. Channels interconnect seamlessly with one another, eliminating the intermediate dead volumes common in macroscopic flow systems that often adversely affect analytical performance. Plus, flowing fluids assume the sample handling and analysis functions previously performed by the analytical scientist, leading to largely hands-off chip operation. The microchip thus also represents an approach to automation of chemical analysis.
Not surprisingly, researchers from many disciplines, but particularly those involved in life science research, have wholeheartedly embraced microfluidics since the mid-1990s as a route to new research tools. The term μTAS was quickly complemented by terms such as microfluidics, integrated chemistry, and lab-on-a-chip - reflecting the broad appeal of the technology.
Landmarks
1979: Miniaturized gas chromatograph fabricated on a 4-inch silicon wafer (9).
1990: Total (chemical) analysis systems (μTAS) systems developed, based on conventional tubing and chromatographic separation technologies (10).
1993: First separation of six fluorescently labelled amino acids in under 15 seconds (11) c1995: First examples of microscale organic synthesis in chips.
Early 2000s: Microfluidics started being used for cell applications 2004: The terms “humans-on-chips” and “organs-on-chip” hit the media.
2007: Development of a microfluidic chip for the isolation and enumeration of circulating tumor cells (CTCs) (12).
2007: First lung-on-a-chip developed (13) 2010: 3D printing made an impact in the fabrication of analytical components.
2010-2012: Microfluidics were deployed in diagnostic tests of infectious diseases in the developing world (14).
2018: Aaron Wheeler and colleagues invented a portable digital microfluidic (DMF) system for use as an immunoassay for testing measles and rubella in remote regions of Kenya (15).
References
- EWK Young et al., Blood 119, E76–E85 (2012). C Pak et al., Integr Biol 7, 643–654 (2015). S Zhang et al., Sens Actuator B-Chem, 263, 614-624 (2018). S Zhang et al., Adv Funct Mater, 1806434 (2018). S Pelt et al., Lab Chip, 17, 3826–3840 (2017). Y Gao et al., Lab Chip, 15, 351–360 (2015). M Humayun et al, Lab Chip 18, 1298–1309 (2018). P Densen, Trans Am Clin Climatol Assoc, 122, 48–58 (2011). SC Terry et al. IEEE Transactions on Electron Devices, 26, 1880-1886 (1979). A Manz et al., Chemical 1, 244-248 (1990). DJ Harrison et al, Science, 261, 895-897 (1993). S Nagrath et al., Nature, 450, 1235–1239 (2007). D Huh et al., Proc Natl Acad Sci USA.,10,18886-18891 (2007). CD Chin, et al., Nat Med, 17, 1015-U138 (2011). AHC Ng et al., Sci Trans Med, 10, eaar6076 (2018).




