Charles Darwin was perhaps the most famous proponent of the importance of the humble earthworm – and they’ve more recently been touted as the most influential species in the history of our world... Unarguably, earthworms play a vital role in our ecosystem – increasing nutrient availability, improving drainage, and contributing to more stable soil structure.
Despite our fascination and nods of approval, methodological challenges have prevented researchers getting a good understanding of the internal workings of an earthworm – including the microbes that colonize it and the associated metabolites they produce. Enter Benedikt Geier and his team at the Max Planck Institute for Marine Microbiology, who set out to accurately image the fundamental interactions between small symbiotic animals and associated microbes in an ecosystem (1). Their recent paper introduces chemo-histo-tomography (or CHEMHIST) – a method for visualizing the metabolic interactions in small animal symbioses, which is based on MALDI-MS imaging and micro-computed X-ray tomography (micro-CT). Their unique approach outperforms similar methods (developed for medical research in mice) by up to two orders of magnitude. We spoke to Benedikt to find out more.
Our team has been working with MS to visually explore how metabolites are distributed in tissues colonized by microbial symbionts. What our research (very quickly) showed us was that we can only understand the chemical images if we know the underlying histology of the host. In other words, we need to know which organs, tissues, and cells of the host animal are associated with symbiotic partners (microbes and parasites) to then assign chemical signals to individual partners.
With this in mind, we set out to create an approach that allowed us to see all this information at once: the host animal’s anatomy in 3D, the associated microbes and parasites that live in the tissues, and most importantly, the metabolites they produce.
Chemical interactions have allowed bacteria to enter symbioses with organisms across all domains of life. These interactions are critical for the health of individuals as well as whole ecosystems – from deep-sea habitats to our own bodies. The problem is, we know next to nothing about the distribution of metabolites and other small molecules in these associations. CHEMIST addresses this knowledge gap by linking the identity of tissues and individual cells in complex tissues to their metabolome – independent of mutualistic or pathogenic associations.
We used an environmental sample – the earthworm – to showcase our technique, because it allowed us to provide a glimpse of the complexity that exists in even the simplest of organisms. Though we have a newfound appreciation for earthworms, our focus is on combining high-resolution MALDI-MSI and micro-CT to enable correlative chemical imaging on a new level. In fact, it was particularly important for us to design an approach that was culture-independent – not only to study symbioses between elusive animals and their microbes, but also to come up with a workflow that would enable the study of human/medical tissue samples. Our approach could also offer new insights into the anatomy and metabolomes of samples that focus on medical studies, such as tissue punctures of pathogen infections.
One major challenge was looking for the associated partners in the host tissues, which can be distributed throughout the whole animal. And that’s not easy when you consider that a small animal host may be measured in 10s of millimeters, whereas bacteria are around one micrometer; the resulting four orders of magnitude in size difference is comparable to looking for a Tic Tac mint in an aircraft carrier!
Therefore, we created a multi-scale 3D imaging approach: first we screen the animal at spatial resolutions just large enough to detect where bacteria or parasites are hidden in the animal tissue, and then we follow up on specific regions with high-resolution MALDI-MSI.
Another challenge was to visualize the broad array of targets from the same animal host: microanatomy to understand the anatomic architecture of the host animal, fluorescence labeling to detect the microbes hidden in the animal’s tissues, and ultimately MS imaging to study what each of the partners produce. Because each molecular imaging tool requires different processing steps of the sample that are not necessarily compatible with each other, we had to find a new way of combining all of them at once.
By analyzing DNA, we include two very important aspects that reach beyond knowing the taxonomy of the associated partners: Firstly, once we know who is there, we can design specific fluorescence probes for FISH to label each species of microbe individually and reveal their community composition within the host tissues. Secondly, because metagenomics sequencing provides information on the genomic potential of the associated partners, we can check “who can do what.” In other words, we can learn which metabolites each partner is capable of producing; by screening the genomes of the associated partners, we can provide essential background information on the metabolite distributions that we record with MALDI-MSI. We described this approach in a paper building up to this publication (2).
MALDI-MSI is extremely powerful because it can deliver both the chemical composition of a sample (like a normal MS) and the spatial distribution of the compounds (like a molecular microscope). On top of that, MALDI-MSI is label-free and allows us to directly observe an animal in its natural habitat, shock-freeze it, and image its body chemistry. As mentioned above, the trickiest part is to understand the chemical images MALDI-MSI produces.
Small symbiotic animals often have complex anatomical structures and even specialized organs or cells that house the symbiotic bacteria – looking in 2D makes it difficult to recognize every structure. This is why we wanted to integrate MALDI-MSI with an approach that would allow us to resolve the detailed 3D anatomy of these tiny host animals at a (nearly) cellular resolution. Although there are fluorescence microscopy techniques capable of 3D imaging, they require tissue clearing approaches for the fluorescence light to illuminate the sample. On the other hand, X-rays pass through any type of biological sample, so we chose micro-CT (an X-ray 3D imaging approach) to resolve and understand the anatomy behind the chemical images that we recorded with MALDI-MSI.
In the MALDI imaging data, we always saw certain molecules that appeared as large blobs towards the end of the earthworm. We could not explain these chemical signals simply from the 2D tissues sections that we recorded. And then we looked at the micro-CT data, which revealed that the blobs were actually cross sections through cysts that contained parasitic nematodes, which had their own specific chemical composition – possibly related to an immune response of the earthworm against these nematodes. This one example showed us that 3D imaging is not only important to understand anatomic and cellular features that might change throughout the animal, but also sheds light on metabolites that indicate certain functions from immune responses (Figure 3 in the paper) to digestive processes along the gut (Figure 1 in the paper).
The diverse set of applications that we envision for CHEMHIST is reflected in the different paths that we have decided to pursue. I am looking forward to taking up my new position as a postdoc at Stanford University, California, where I will transfer my knowledge in correlative chemical 3D imaging to organoid-infection models to better understand human pathogenesis through biomedical research. Manuel Liebeke, one of the key members of our team, is Head of the Metabolic Interactions Group at the MPI for Marine Microbiology in Bremen, Germany, and just recently received a new MALDI imaging setup for faster and higher resolution imaging, which he will use to further push the boundaries of imaging symbiotic interactions. In particular, he will be using a specific MALDI-MSI workflow that enables imaging of glycans – cell surface molecules that are involved in the metabolic interactions at the host-microbe interface.
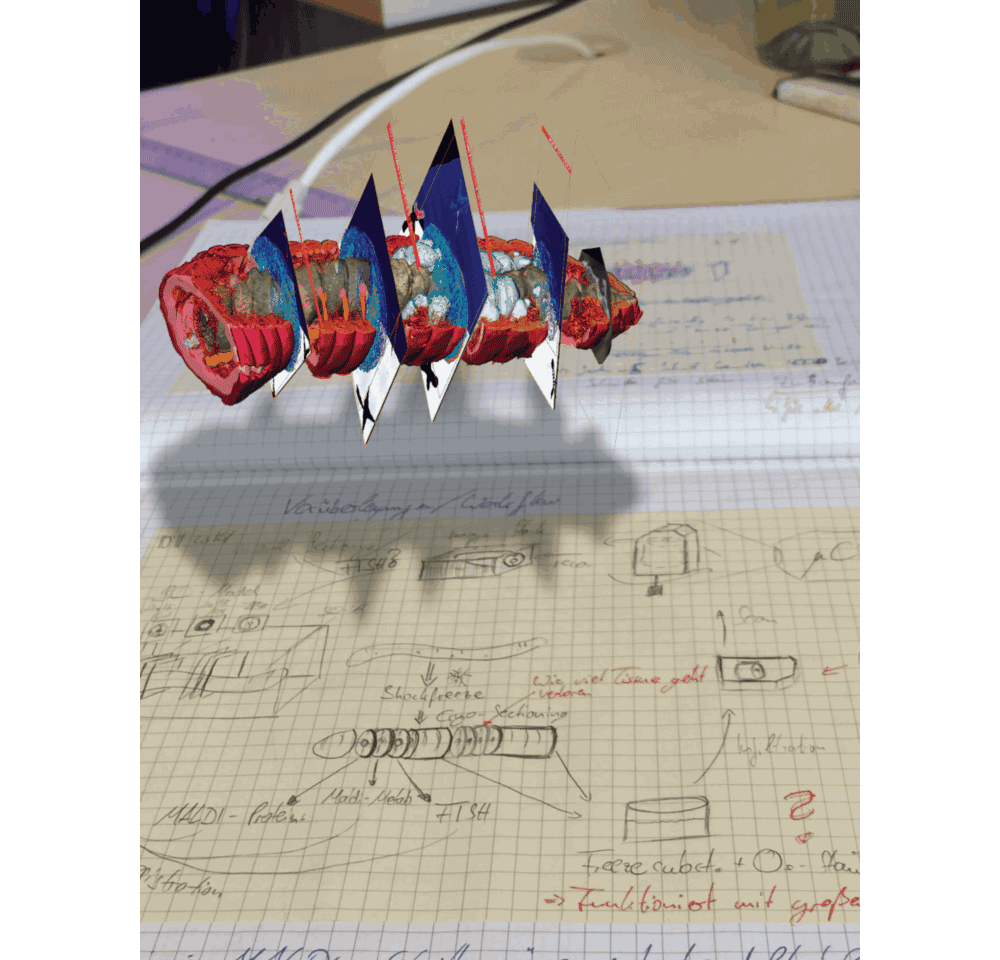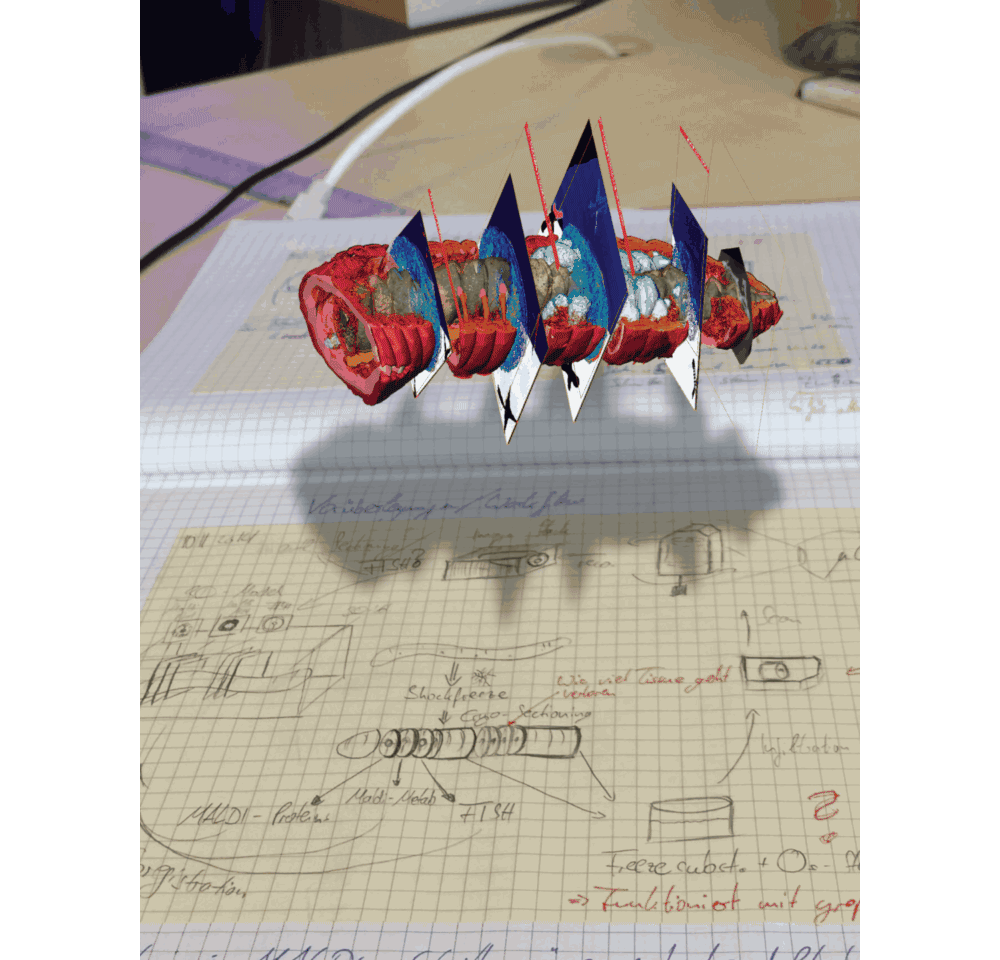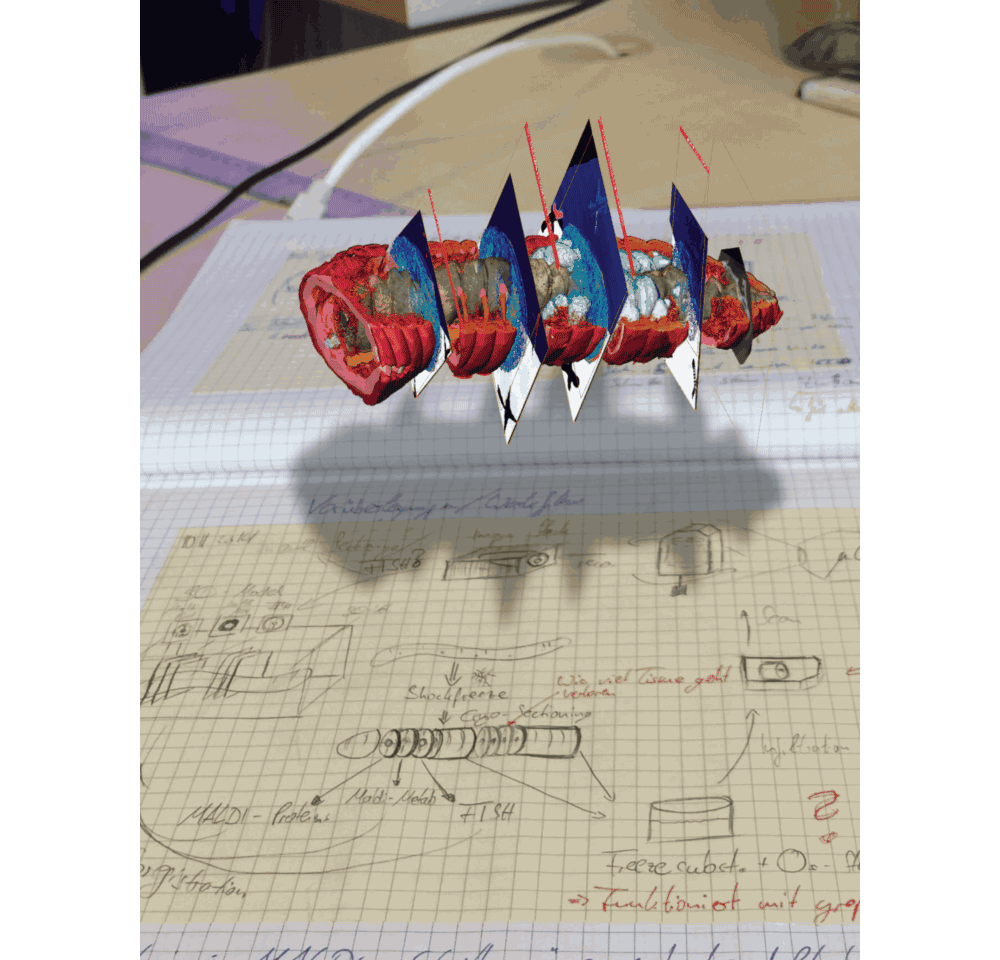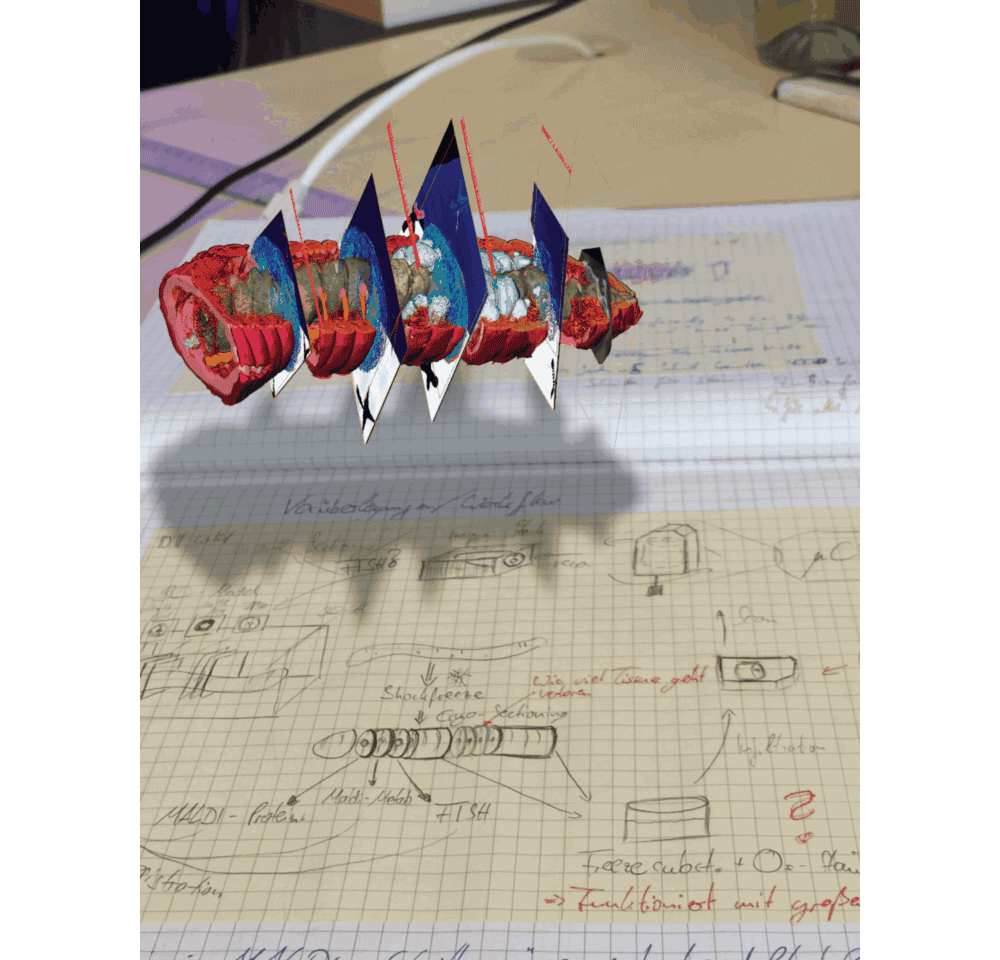
We are convinced that correlative chemical imaging – as presented in our CHEMHIST study – will lead to more work on host-microbe interactions. In terms of a technical outlook, the increasing speeds, spatial resolutions, and sensitivities of imaging setups will allow scientists in the future to not only create a multimodal 3D atlas of one animal, but maybe even a whole time series of different infection or life stages. CHEMHIST is designed as a framework that future scientists can adapt to their needs for studying any symbiotic/host-microbe system.
We would love to see CHEMHIST applied as broadly as possible! For example, to specific medical questions where scientists want to know what metabolites the pathogens, parasites, or maybe even beneficial microbes (for example, in the gut microbiome) produce within their host niche.
One aspect in particular that we wanted our study to highlight is the value of discovery-driven research; for example, observing a system from a different angle and generating hypotheses on the interactions taking place in front of one’s eyes. When Darwin studied the Galapagos Islands, he would first observe and describe what he saw before trying to explain it. In a similar way, seeing the distribution of metabolites and the animal and bacterial cells that produce them embedded in the anatomic 3D “world” of the host’s body is like discovering a micrometer-scale ecosystem. New imaging methods enable new observations – similar to discovering an unknown island – which allows us to first observe nature and from there create hypotheses that we can test, discuss, and begin to explain. As Thomas Bosch said in his commentary on our work (3): “How excited Charles Darwin would have been if he had found out that the earthworm’s behavior might be the result of such complex multiorganismic interactions.”
References
- B Geier et al., “Connecting structure and function from organisms to molecules in small-animal symbioses through chemo-histo-tomography,” PNAS, 118, e2023773118 (2021). DOI: 10.1073/pnas.2023773118
- B Geier et al., “Spatial metabolomics of in situ host–microbe interactions at the micrometre scale,” Nat Microbiol, 5, 498 (2020). DOI: 10.1038/s41564-019-0664-6
- T Bosch, “Taking a microscale look at symbiotic interactions—and why it matters,” PNAS, 118, e2110874118 (2021). DOI: 10.1073/pnas.2110874118




