Thallium sulfate has no taste, smell or color but is highly toxic – and that’s probably why it goes by the name of The Poisoner’s Poison in certain circles. “I remember about twenty years ago there was a well-known murder case in China in which thallium was used as a poison – the mystery remains unsolved even today,” says Juewen Liu, assistant professor at the University of Waterloo, Canada. Liu wondered about the potential of a thallium biosensor, but could find very little in the literature. “It seems strange that its two neighbors in the periodic table, mercury and lead, have been so extensively studied, but not thallium,” says Liu, who has focused on developing DNA-based ligands for metal detection for the past 15 years. To redress the imbalance, Liu and colleagues decided to develop a method that can indirectly detect Tl3+ using a catalytically-active DNA molecule, known as a DNAzyme.
DNA is a highly stable, programmable, and versatile component in the design of sensors. And compared to current instrumental analysis methods, DNA-based biosensors are more cost-effective and portable. The big problem is that Tl3+ has very little interaction with DNA, which likely explains the lack of research, says Liu, who found a workaround. The new sensor uses a DNAzyme called Tm7 isolated in recent work (2). Tm7 is an Er3+-dependent RNA-cleaving DNAzyme with an unusual property. “We discovered that if we introduce a phosphorothioate (PS) modification into the RNA substrate, Tm7 becomes completely inactive – it cleaves only the normal phosphate (PO) RNA substrate,” explains Liu. “This is the first DNAzyme ever reported with this property. For most other DNAzymes, partial activity is retained even with the PS-modified substrate.”
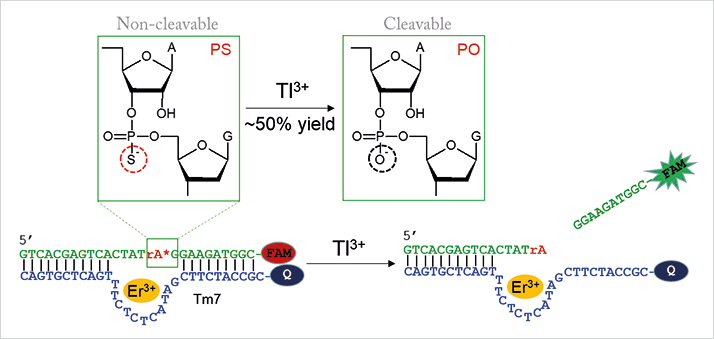
What does this have to do with thallium? In the presence of Tl3+, which is strongly thiophilic, the RNA linkage is desulfurized, meaning that it can subsequently be cleaved by the DNAzyme (see Figure 1). The RNA substrate is labeled with a fluorophore that is quenched when bound to the DNAzyme. The cleavage reaction releases the fluorescent fragment and therefore results in Tl3+ concentration-dependent fluorescence enhancement. Liu notes that other approaches, such as colorimetric or electrochemical detection, may also be possible. Liu adds that though Hg2+ and other thiophilic metals are able to desulfurize the RNA substrate, they also inhibit Tm7 – Tl3+ does not inhibit the DNAzyme, which offers some specificity. And according to Liu, sensitivity goes down to 1.5 nM – below the maximal contamination limit defined by the US Environmental Protection Agency (10 nM). One current limitation is that the sensor can only detect Tl3+ and not Tl+. “We are now trying to develop a Tl+ sensor using DNA and hope to use both sensors to study metal speciation. We are also interested in finding the difference between Tl3+ and Hg2+ to allow better separation of these two metals,” adds Liu.
References
- PJJ Huang et al., Desulfurization activated phosphorothioate DNAzyme for the detection of thallium”, Anal Chem, 87, 10443-9, (2015). DOI: 10.1021/acs.analchem.5b02568. PJJ Huang et al., “A new heavy lanthanide-dependent DNAzyme displaying strong metal cooperativity and unrescuable phosphorothioate effect”, Nucleic Acids Res, 43, 461-469 (2015). DOI: 10.1093/nar/gku1296.




