The importance of the global pharma industry has rarely been more evident. Though much of the work happens behind closed (high security) doors, exciting advances are transforming the way we can create and study drug molecules – from inception to injection.
Inspired in part by the attractive pictures they produce, we set out to explore the application of modern analytical techniques to imaging drugs, not just in the body or tissues, but at the subcellular level.
Please welcome Michael Kurczy and Alison Hulme, leading minds from the realms of MS imaging (MSI) and spectroscopy, respectively, who were kind enough to let us pick their brains on the matter! Now let’s get to the interviews…
Our main focus is to design a reliable method to localize and quantify drug candidates at the subcellular level. We are developing our platform using a Cameca NanoSIMS 50L, so I feel the need to mention how unique our situation is. We work in an incredibly collaborative environment as part of the Chemical Imaging Infrastructure (CII) at AstraZeneca, which is supported by Gothenburg University and The Chalmers University of Technology. These institutes are in turn hosted by the AstraZeneca BioVentureHub – an open innovation framework that makes infrastructure operation possible at our Gothenburg site. In addition to researchers at AstraZeneca and the CII, we have also had postdocs funded by the AstraZeneca postoc program and an industrial PhD student working on this endeavor.

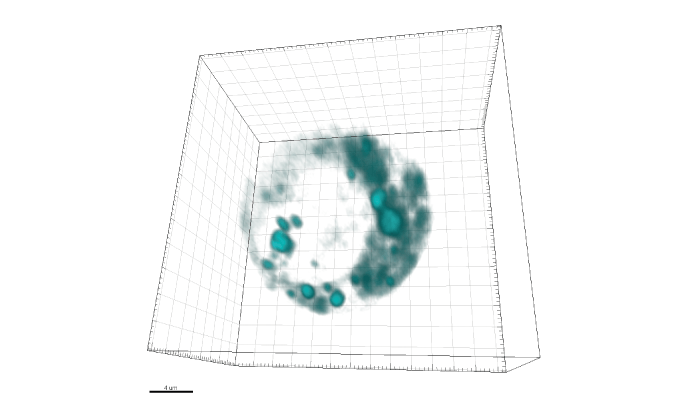
Because that is where the action is! As we say in the paper, the cell may be the basic unit of life, but disease pathways are regulated at the subcellular level. The real question is thus: “how well does a measurement at the plasma or tissue level reflect the situation inside a cell?” Our approach analyzes individual cells rather – not a tissue homogenate. We knew that the spatial resolution of the NanoSIMS was adequate to image drugs at the subcellular level, but we felt driven to report absolute concentrations to give meaning to the resulting data. We spent a lot of time validating the idea that we could translate isotopic ratios into the concentrations of isotopically labeled drugs.
The idea of transforming an isotopic ratio to a concentration is certainly a new concept. It’s a strange idea – and one that would only come from collaboration between a bioanalytical chemist and a geologist. People conducting biological SIMS don’t typically report limits of detection (LODs), but geologists do! We wanted to understand our LOD in more depth, and – luckily – our facility manager (and lead author on the resulting paper) Aurélien Thomen has a geology background.
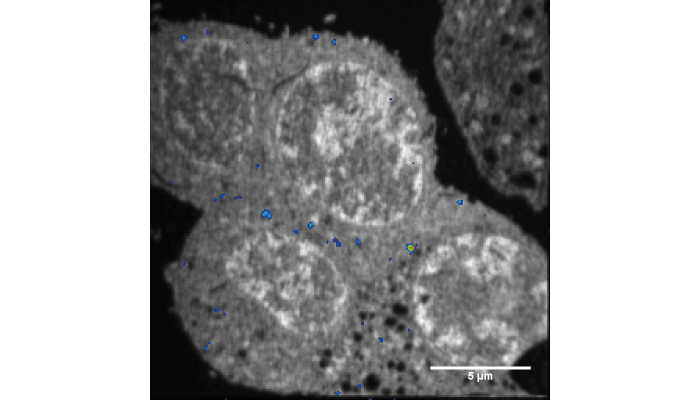
We decided to treat biological samples as homogeneous material. This works well because, when we analyze cells with NanoSIMS, they are embedded in an epoxy that replaces the intracellular water. All we had to do then was calibrate the isotopic enrichment of 13C to the carbon density of the epoxy. This was fairly easy to calculate, and easy to test!
But we also needed a real cell sample with a known concentration of an isotopically labeled compound. There are not many examples of such a system. Fortunately, Andrew Ewing (Gothenburg University and Director of the CII) had characterized the dopamine concentration contained in exocytotic vesicles in rat pheochromocytoma cells (PC12) – just what we needed! It was somewhat straightforward to load these vesicles with 13C-labeled dopamine. In fact, we jokingly call this sample the Rosetta Stone, because it allowed us to confirm the translation of isotopic ratios into concentrations.
This is really the first step, but we have now shown that we can speak the language of DMPK (drug metabolism and pharmacokinetics). Researchers can use bioanalysis to measure drug concentrations in plasma or tissue to create a profile of concentration over time. This approach is used to model how a drug moves though the body. That’s where our research comes in. We anticipate that adding a subcellular component to these kinds of models will be possible in the near future. And the result? Increased knowledge and an accelerated drug discovery process.
It’s my dream that this technology become a routine and mainstream method – every pharma company could have a NanoSIMS in the lab! At that point, we hope to have a suite of standards to cover an array of labeling strategies, as well as standardized sample prep protocols and instrument parameters. But the biggest hope is that people working in pharma will be comfortable collecting and understanding NanoSIMS data and using it to make decisions that help us develop effective therapies more quickly.
The CAMECA NanoSIMS is a big investment, so its biggest obstacle is access. We were lucky enough to use a machine that had been purchased by a nearby university – maybe collaborative frameworks like this will be important for improving access. But the key, as I said before, will be making those working in pharma comfortable with the data that this approach produces. I believe that reporting an absolute concentration is a big step in that direction. As things stand, however, we're the only company with this instrument available in one of our buildings! We’re privileged in this way. Best of all, it means that we have a lot of time to test and learn, and to create new methods!
I trained as a synthetic organic chemist, but I was never entirely happy with just making molecules. I have always been fascinated by the biology of the molecules I make, as well as their chemistry – an interest that found its roots in my mother (a biology teacher). She would bring high school experiments home to show us, such as dogfish (very stinky!) and cheek cells under a microscope – fascinating! My group’s research integrates synthetic methodology development with chemical biology to provide molecular-level insight into challenges in biology and medicine. I collaborate with academics working in the fundamental biosciences right through to hospital clinicians.
We use the standard range of analytical tools for synthetic chemistry, including LC for purification, and NMR and MS for characterization of the compounds we make. Over the past seven years, we have combined our synthetic expertise with that of Val Brunton (Professor of Cancer Therapeutics at the Cancer Research UK Edinburgh Centre) and Martin Lee (Institute of Genetics and Molecular Medicine Advanced Imaging Resource, University of Edinburgh) to develop an emerging technology, stimulated Raman scattering (SRS) microscopy, for biomedical applications.

Leica have just introduced the first SRS microscopes to the market, but, not wishing to boast, these commercial instruments aren’t quite able to produce the resolution or quality of image that we can produce in the laboratory – after all, we are able to control all the component parts. People are now exploring applications for the instruments, for example, in histopathology, around the world – largely in the UK, US, Germany, and Japan – but the number of these microscopes in routine use is still somewhat limited. Our aim is to encourage uptake of the instrumentation by showcasing its potential in biomedical applications.
Our research in this space has focused on imaging drugs that have an inherent vibrational motif in the cell silent region of the Raman vibrational spectrum, as well as developing new tags that can track drugs lacking these inherent vibrational features. In both cases, we have focused on alkyne motifs because these give spectral peaks that are approximately 100 times narrower than fluorescence peaks, and occur in a spectral region with low background noise. We image the cellular structure and the drugs by using the inherent vibrational spectra of biomolecules (such as proteins and lipids), which can be obtained without any fixing or staining. The result: live-cell studies!
Our team has acquired some of the highest resolution live-cell images of intracellular drug distribution to date. A great example is our 2020 study published in the Journal of Medicinal Chemistry (2), in which we used SRS microscopy to image the tyrosine kinase inhibitor ponatinib at biologically relevant concentrations without labeling. This study also provided insight into changes in uptake and sequestration of the drug that occur during the development of drug resistance – informing us how the drug worked initially, and the mechanisms by which its function was interrupted. These promising results have placed the field of SRS microscopy firmly in view of the pharmaceutical industry as a means to improve drug development.
Despite the identification of an unprecedented number of potential new drug targets over the past two decades, and accompanying investment in the generation of new chemical entities with improved potency and selectivity, only 10 percent of clinical candidates progress to regulatory approval. Incorporating imaging into complex drug screening models has the potential to improve the robustness of preclinical studies of drug uptake, retention and metabolism. The insight gained from such studies could enable earlier removal of ill-fated compounds from the development cycle, leading to improved drug quality and lower financial risk.
Though there are a number of imaging methods available for the analysis of drug uptake, distribution, metabolism and mechanism of action, Raman presents with a number of advantageous traits. An example: the lack of need for external labels to image cellular structure, as Raman relies on vibrational frequencies in functional groups! Raman also exhibits higher spatial resolution and weak water interference, which makes it suitable for biological applications. Other imaging methods, such as PET and MRI, suffer from low spatial resolution, and MS requires extensive sample preparation – limiting our ability to image in live samples.
There are limitations in our current capabilities too, though. For example, even with these coherent Raman microscopy techniques, we obtain comparatively weak signals, meaning we rely on relatively high local intensities to obtain our results.
One area that has been being explored, but not really exploited yet, is the use of multiple spectral frequencies simultaneously. I expect that we will see more of this multiplexing in the future – as well as a greater focus on this technique (and other imaging techniques) in general. The pharma industry (and AstraZeneca and GlaxoSmithKline in particular) is already becoming increasingly involved in imaging networks that will allow them to capitalize on these emerging technologies.
I also anticipate that current challenges, such as the need to develop vibrationally intense chemical tags to amplify signals that are also metabolically stable and exert minimal impact on the drug being studied, will be overcome in innovative and ingenious ways. And a last exciting prospect is the development of approaches that incorporate multiple imaging modalities; these will allow researchers to study drug effects at multiple levels, from the whole body down to specific tissues, cells, and organelles.
References
- A Thomen et al.,”Subcellular mass spectrometry imaging and absolute quantitative analysis across organelles,” ACS Nano, 14, 4316 (2020). DOI: 10.1021/acsnano.9b09804
- K. Sepp et al. “Utilizing stimulated Raman scattering microscopy to study intracellular distribution of label-free ponatinib in live cells”, J. Med. Chem., 63, 2028 (2020) DOI: 10.1021/acs.jmedchem.9b01546




