I didn’t set out to work in 3D printing. In fact, when I applied to do a PhD with Conan Fee and Simone Dimartino at Canterbury University, New Zealand, it was on a completely different topic. But in the eight months it took for me to receive my New Zealand visa, Conan and Simone had an idea about the capillary-polymer phase fiber columns they were working on: why not just 3D print the fibers exactly where they are needed? In fact, why stop at fibers? Why not just print perfectly ordered particles and open up all manner of possibilities for liquid chromatography? When I finally made it to New Zealand, I was given a choice: either work on the original topic that I applied for – or take on this new area of 3D printing. I was so struck by the potential of the 3D printing approach that I immediately decided to pursue it, despite knowing virtually nothing about 3D printing... or liquid chromatography!
In perfect order
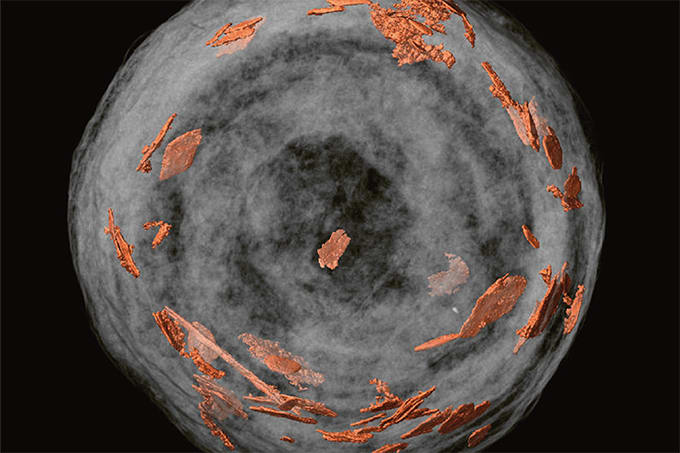
With traditional slurry-packed LC columns, random placement of particles hinders the column’s performance. It is possible to create monolithic structures, as Frantisek Svec (E.O. Lawrence Berkeley National Laboratory) and Sebastiaan Eeltink (Vrije Universiteit Brussel) have done. Still, there remains the issue of the random placement of microglobules that are cross-linked. In either case, we end up with column geometry that exhibits inherent randomness. Our initial idea was to make a completely ordered lattice of particles; there are plenty of computational studies that showed the advantages of uniform packings, so our goal was to replicate them experimentally. Another good feature of 3D printing is that we can design and make fittings, distributors and column walls, all in one piece – they are very much plug-and-play. So far, so good – but turning theory into reality often requires answers to a number of questions. What are the characteristic dimensions? What are the geometries we want to design? What materials should we use and how do we get from a printed structure to a fully functionalized column? Can we use printers that exist in the market or do we need to make our own? Each of these questions is a PhD project in its own right... I decided to focus on the use of inert plastic models, such as the one shown in Figure 1, to create structures that could replace packed beds. Essentially, I sought to answer the question: what is the best stationary phase geometry for LC? But it wasn’t just about designing the packing structure – we could also change the flow distributors and column shapes, and we found that it had a significant influence on column performance (1).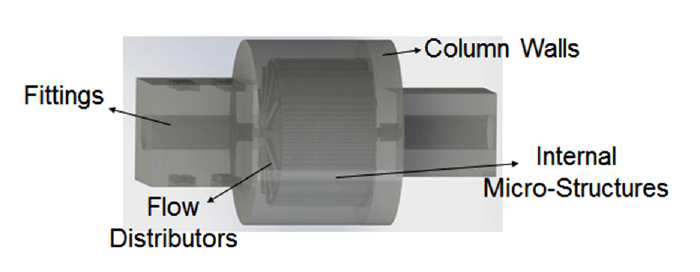
A rare geometry
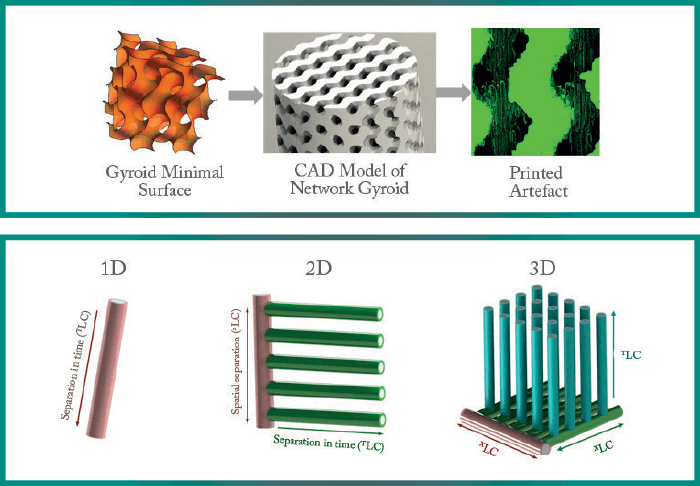
We began by making columns with uniformly stacked ‘particles’. The particles slightly overlap so they are monolithic structures in the strict sense of the word. We can control the size, shape, arrangement and orientation of these ‘particles’. We showed that arranging spherical particles in a face-centered cubic formation is the optimal set-up. And though this fact has been shown many times in computational studies, it was exciting to confirm the theory experimentally for the first time! One result that none of us saw coming was that tetrahedral ‘particles’ outperform spherical particles when arranged in ordered lattices (2). We don’t need to limit ourselves to packed beds with particle-like elements. Mathematicians have spent whole careers coming up with and studying all sorts of interesting geometries but, until now, there had been no way of physically making them. With 3D printing, that obstacle largely disappears, and suddenly we have access to the best geometries that mathematics has to offer. For example, minimal surfaces – soap-film-like sheets designed to minimize surface tension, resulting in warped, curved geometries, such as gyroids (Figure 2). They have been explored computationally but thanks to 3D printing we could confirm that minimal surfaces beat the best sphere packings by a factor of 6-8 when it comes to separation impedance. We are only just scratching the surface in terms of the geometries we can build. Minimal surfaces are just one family of mathematically interesting structures; there are many more that we can and should study. Then, we can answer the question of what the perfect stationary phase structure for LC really is.
3D=Freedom
One of the things that makes working with 3D printing so enjoyable is the speed with which we can produce alternative designs – we can go from an “a-ha” moment to a 3D design in an hour and have the new part printed overnight. Next morning, we’re ready to test the new part and see if it does any better. An example of this is flow distribution. The goal is to achieve uniform flow from a single point (the tip of the inlet tube) to the cross-section of the column. Our first design fell down on this point; we couldn’t even tell the difference between different packing geometries. We were able to rapidly change tack, and come up with a new flow distributor design based on a Manelbrot H-tree fractal, which was much more successful.STAMPing out slow separations
My PhD supervisor Simone Dimartino presented our group’s work at HPLC 2016 in San Francisco. Peter Schoenmakers and his team from the University of Amsterdam were also there. And, as part of their work on 3D separations, they were looking for someone with 3D printing experience to join their exciting STAMP (Separation Technologies for a Million Peaks) project; Simone recommended me, and I set off on a new adventure. As everyone in the field knows, one of the main challenges in liquid chromatography is the need to increase separation of very complex samples. Right now, we’re several orders of magnitude short of the peak capacities we need for proteomics and metabolomics. The number of peaks that a single column can resolve has improved over the years, but with steadily diminishing returns. We can make longer columns with smaller and smaller particles but eventually it turns into what Schoenmakers calls “ultra-high patience liquid chromatography” – higher and higher backpressures and very long analysis times. In the late 1970s, 2D-LC came onto the scene. Fractions from the first column are injected onto a second column with a different retention mechanism, dramatically increasing the resolving power of our overall systems. In principle, 3D separations are no more than an extension (or added dimension!) of 2D-LC; the goal is the same – to achieve greater peak-capacities. That said, with 2D-LC, we typically think of a coupled-column technique, where fractions from the first dimension separation are transferred to the second. The drawback is that the analysis times are long, because fractions of the first dimension are analyzed sequentially in the second. The alternative? Performing the separations in parallel rather than in series. For this, the first dimension has to be a separation in space. Fractions are also in space rather than time. We can have several perpendicular 2D columns attached to the 1D. This way, these spatial 1D fractions can be transferred to the second simultaneously, and we dramatically reduce the total analysis times. STAMP aims to achieve radically high peak capacities at short analysis times by carrying out separations in parallel rather than in series; separation in space rather than time.Three really is the magic number
As early as 1983, Georges Guiochon et al. described a device for “development in the first two dimensions and elution in the third” – our main working principle (3). They envisioned the device as a single 12x12x12 cm box, filled with particles, where the three separations would occur. Thirty-four years later, we’re still trying to make those principles a reality! Why has it proved so challenging? For a 1D separation, the ideal shape is a single line; the closest we can get is a long, thin cylinder. For 2D parallel separations, the first dimension has to be a spatial separation, which branches off into several second dimension channels. For this, a planar device is ideal. When we go to 3D, the first two dimensions need to be spatial separations. Bert Wouters has made a first prototype that works in this manner during his PhD research at the Vrije University Brussel and now works with us on STAMP. As you can see in Figure 3, when we add more dimensions to our separations, the geometries become very complex very quickly! Effectively, we’re trying to house hundreds or even thousands of mini-columns, complete with valves and distributors in one device. We need to transfer the analytes from the first to the second to the third column in a very controlled manner. And we need to do this without transferring the mobile phases of each dimension to the next. Needless to say, any device of this kind will need to have an incredibly complex structure. Part of what we do at STAMP is to design and optimize these devices. It might be possible to create these devices with the same methods we use for microfluidic chips (micro-milling, PDMS replica molding, and so on) but there are limitations in terms of complexity. And that’s where 3D printing comes into its own: transforming the impossible into simply very difficult. With 3D printing, there is no upper limit to the complexity of parts we want to build. For me, it’s been an exciting opportunity to work on a ‘moonshot’ project and put my knowledge on 3D printing to good use. The main application that we’re targeting is proteomics and metabolomics, where extremely high peak capacities are necessary prior to mass spectrometry (MS). For a good MS analysis, you cannot have dozens of co-eluting compounds with a large range in concentrations.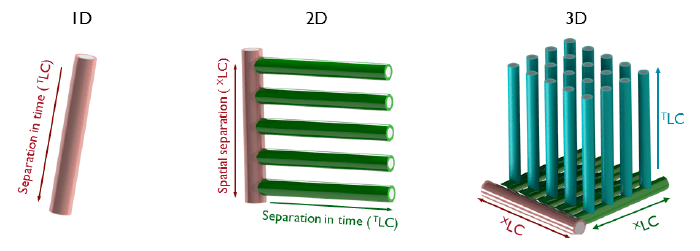
The technology still has a long way to go. As well as the fluid mechanical challenge, there is the difficulty of finding a combination of retention mechanisms that are both compatible and orthogonal. Then, there’s the small matter of detection from dozens of eluent streams in parallel. This is why we have five PhD students working on solving different parts of the jigsaw puzzle. These challenges mean that we likely won’t see 3D-LC in use for several years. It took column-based 2D-LC a few decades to go from a concept, into academia and finally into industry. We have the advantage that we can piggyback on much of the research that has been performed on column-based 2D-LC – and in the next decade I hope to see 3D devices beginning to appear in university laboratories for proteomics research. Add another decade or so and we can expect it to seep into commercial research facilities. Suhas Nawada is a postdoctoral researcher in the STAMP project (Separation Technology for a Million Peaks) at the University of Amsterdam’s Van’t Hoff Institute of Molecular Sciences, the Netherlands.
References
- C Fee, S Nawada, S Dimartino, “3D printed porous media columns with fine control of column packing morphology”, J Chromatogr A, 1333, 18–24 (2014). G Guiochon et al., “Chromatography with a two-dimensional column”, Chromatographia, 17, 121 (1983). B Wouters et al., “Towards ultra-high peak capacities and peak-production rates using spatial three-dimensional liquid chromatography”, Lab Chip, 15, 4415–4422 (2015).