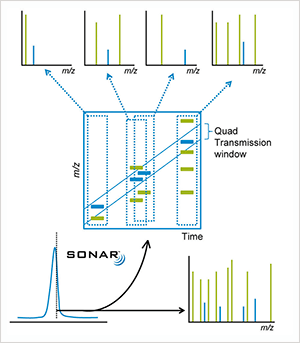
With John Chipperfield, Senior Systems Evaluation Scientist at Waters. SONAR is an elegant solution to a very specific problem; if you’ve got high complexity in your sample and you need more reliable information, then this is the mode for you. SONAR is a data independent acquisition mode that generates MS/MS data – at UPLC speeds – for everything in the sample indiscriminately, using a resolving quadrupole that scans rapidly over a given mass range. Essentially, SONAR enables users to quantify and identify components from a single injection. It’s actually a pretty straightforward implementation of a simple idea – it just so happens that it’s very powerful. For users to be able to stay ahead of the game, they need to be able to achieve more with less time – and that means gaining as much information from a single analysis as possible. But there’s an important point to make here: speed is nothing without confidence and reliability. Analysts need to be able to trust their data – they need to know that they have correctly identified a given compound – and that’s a crucial issue that we aimed to address with the SONAR acquisition mode. SONAR is an incredibly powerful qualitative technique that can generate a tremendous amount of information for a given sample – and it does this faster and much more thoroughly than you could before. But, to reiterate, you can also pull quantitative information straight out of that dataset. Importantly, it’s just as easy to process SONAR data as any other data that we produce – you can just plug it into Progenesis or UNIFI. And having listened to our customers, we’ve also made it compatible with third-party software – Skyline from the MacCoss lab, for example. If we compare SONAR with our major competitor in the area of data independent acquisition, there’s an important distinction to make: because we can scan so fast using a resolving quadrupole, we gain selectivity and we’re able to assign fragments to precursors very accurately. But we’re also able to do it quickly enough to get the optimum number of points across a peak to enable quantification. I don’t think our competitors can match up on that particular aspect.
SONAR Product video with Jim & John:
With Jim Langridge, Director of Scientific Operations at Waters What’s your background? I started out in biochemistry and analytical chemistry. While I was working on traditional biochemistry techniques – ELISA and fluorescence assays, and so on – we began to explore the potential of mass spectrometry to replace some of these methods. The technology was going through something of a revolution at the time and seemed to be accelerating faster than traditional biochemistry. So that was my route into mass spectrometry. I’ve been with Waters for 23 years, taking on a variety of different roles during that time; I worked on the original Q-TOF that we developed in 1997, and also on the early incarnations of the ion mobility technology that we’ve pioneered in the SYNAPT platform. I’ve also spent a lot of time looking at protein and peptide analysis using chromatography and mass spectrometry, and more recently metabolite and lipid analysis, as these are becoming increasingly important. I’m now Director of Scientific Operations at Waters in Wilmslow, but also hold an honorary professorship at Imperial College London in the Department of Surgery and Cancer. The latter role came about through my involvement in projects on desorption electrospray ionization (DESI) and rapid evaporative ionization mass spectrometry (REIMS) technology, developed by Professor Zoltan Takats, who works with Professor Jeremy Nicolson at Imperial. You’ve seen more than a couple of decades of mass spectrometry development – how have you seen the field change during that time? If you go back 25 or 30 years, most people were interested in investigating single analytes that held specific biochemical significance. I think the biggest change that we’ve seen is the drive towards profiling a large number of analytes – almost taking a snapshot of a particular biological process by investigating the proteins and underlying metabolites. Today’s researchers want to use this information to derive biological context – mapping pathways and interactions to understanding how that relates to a disease state, for example. Such analysis has only been made possible by advances in electronics and computers. As power and speed have increased, we’re now able to scan faster, with better sensitivity, than I could have imagined – and that’s opened up huge future opportunities in biology and health. So we need to look at the bigger picture? Exactly. And you can see how research is already starting to change. In the future, people will be more interested in flux-based studies, looking at how molecules and systems change over time. That’s part of the reason we developed our SONAR technology – to give researchers a tool that not only offers very powerful qualitative information (something that allows you to get specific information for metabolite identification), but also quantitative information, which is essential for flux studies and understanding how analyte concentrations are changing between different sample sets. At Waters, we innovate by taking a slightly different pathway – and in doing so, we are able to bring new capabilities – such as SONAR – to the market. That’s probably why I’ve stayed with the company for over 20 years... SONAR is the latest in a very long line of exciting developments that, for me, started with the SYNAPT platform, and includes REIMS and DESI. It’s a great time to be in science and it’s a great time to be at Waters. What do you consider to be the key challenges facing modern research? Perhaps the most relevant word is “complexity” – especially in terms of sheer numbers of analytes in a given sample. Dynamic range also comes into play; very complex samples tend to include analytes over a very wide dynamic range, which can put stress on typical approaches, and cause potentially important analytes to be missed. Even today, we can’t see all analytes, but each iteration of technological improvement peels away another layer of complexity, allowing us to see many more components than before. Speed and sample throughput is also an issue – with high complexity comes the need for more rigorous results, which demands analysis of an ever-higher number of samples. Gaining robust data across large numbers of samples is essential if we are ever to understand the underlying biology. Without such an approach, we risk making all sorts of assumptions that aren’t valid. I’m pleased to say that this field of research seems to be moving away from studies with limited samples. Savvy researchers realize that you need to analyze a large number of samples to find something significant. It turns out biology is a lot more complicated than we once thought! Determining the (changing) concentrations of many analytes in large numbers of complex samples is hugely challenging. But as technology advances, researchers are able to do much better and more comprehensive studies.
SONAR: An expert’s perspective
What makes SONAR special? Data independent analysis has been around for over ten years – driven by the need for unbiased approaches that don’t rely on making decisions on which ions to sample or fragment, but instead record information on all ions. SONAR has two advantages: first, the scanning nature of the quadrupole gives us increased specificity of what we’re selecting; because we have a resolving quad, we know what we are passing through at any given point in time, which gives us selectivity – useful when you’re trying to associate fragment ions to precursor ions in qualitative work. The second interesting aspect – and what’s different about SONAR – is that we don’t need the quadrupole to be at a very high resolution; instead we can actually de-resolve it, transmitting a wider window to effectively gain in sensitivity. Those two aspects give us a very flexible acquisition mode, which gains its true power through informatics processing. How does SONAR address the throughput issue? The fact that modern electronics now allow us to scan quadrupoles at 10,000 amu per second means that we can move the quad extremely quickly, which, in practical terms, allows us to collect more than ten spectra per second across a chromatographic peak. Consider modern separation science, where the peaks are getting narrower either because people are trying to gain better resolution or increase throughput. The number of points we get across that peak is important both from a qualitative and quantitative perspective. You mentioned the power of informatics processing – could you go into more detail? One of the big changes we’ve made in terms of company philosophy is our decision to open up our software environment so that people can access the data and use them in a variety of different software packages. We had a big push at ASMS 2017 on that very subject. We’ve worked – particularly in the research environment – with a wide variety of scientists who have different requirements. They wish to look at data in different ways and use different programs and open source tools. The community has definitely driven our decision, but we also recognized that if someone uses an open source tool to get the information and the results they need, our facilitating that process can only be good for us and our customers. We realize that we cannot provide every single aspect of functionality demanded by such a broad spectrum of different applications and requests. Skyline came out of MacCoss Lab at the University of Washington, and when I first saw it, I remember thinking, “Wow! What a neat approach. This guy has some great ideas.” At an early stage, when Skyline was struggling to gain traction, we were very supportive and worked hard to get our software to produce data in the right format. Interestingly, our effort also encouraged other vendors...
SONAR is a simple concept – why has it not been done before? It is simple, but was only made possible by the work we’d done to access data independent acquisition through ion mobility on the SYNAPT platform, which demanded the development of a novel acquisition system that could acquire up to 2,000 spectra per second. For the XEVO and SONAR, we basically used the same acquisition system but, instead of using ion mobility, we used a scanning quad in front of the time-of-flight, which allows us to store data in 200 bins per quad scan. In other words, the idea is simple, but the execution was only made possible by a very talented development team! Who’s most excited by the potential of SONAR? Definitely the lipidomics community, where we feel there is a big opportunity in terms of an unmet need, namely increased selectivity. Lipids are very close in mass-to-charge, and there are many structural isomers. SONAR allows you to do one acquisition, but still pull those apart quite nicely. Proteomics is another big area. We’re also starting to see it move into other areas as we further develop the technique; the mass spectrometry imaging community, for example. We actually had a presentation at ASMS 2017 based on the use of SONAR with DESI imaging. What about outside of health science? Environmental analysis and foodomics is a natural progression. Like most technologies, when we first develop them we apply them to the most challenging application area – in this case, proteomics, metabolomics, and lipidomics. But challenges are often mirrored in other fields. Another area that we’re starting to explore is biopharmaceutical characterization. Here, reliability and reproducibility of results is absolutely critical, which plays to the key strength of SONAR.
Download SONAR Whitepaper