

Chronic ulcerative wounds are a distressing and common complication of diabetes. To help determine the processes that prevent normal wound repair in diabetic patients, the McLean Research Group based at Vanderbilt University, Tennessee, used ion mobility-mass spectrometry (IM-MS) to analyze wound fluid (a complex brew of byproducts of the wound repair process) from sponges implanted into the wounds of diabetic and control rats. Here, Kelly M. Hines, lead author of the group’s recent paper (1) answers questions about the research.
The ion mobility-mass spectrometry (IM-MS) approach keeps the wound fluid intact. This allows a broad scale and untargeted investigation of the interactions between multiple types of molecules involved in the wound healing process. The approach spans the transition from untargeted to targeted analysis: differentially expressed molecules are revealed in the untargeted data set, while the guidance of statistical analysis and further experiments are performed in a targeted fashion to elucidate and validate their identities.
It was interesting to observe biomolecular species in wound fluid that are similarly differentially expressed in the serum and blood of diabetics. Lysophoshatidyl choline (20:4), cholic acid and the protein S100A8 were associated with diabetes in previous studies, while S100A8 has been implicated in the wound healing process and diabetes.
For the initial untargeted analysis, electrospray ionization (ESI) was used, and ultra-performance liquid chromatography (UPLC) was introduced for the subsequent targeted analysis. The separation of multiple classes of biomolecular species in the IM dimension is greatly beneficial for the analysis of complex samples because it allows us to integrate various omics analyses in the same experiment with very little sample preparation. For example, it allows lipidomics, metabolomics, and proteomics to be performed simultaneously. Omics analyses by MS only typically requires extensive sample preparation to deplete the undesired species from the biological samples, reducing the native complexity of the sample. Using IM-MS, sample preparation procedures were reduced to dilution and desalting (for ESI). UPLC analysis was performed in a targeted manner to elucidate the source of the molecular species m/z 355 and 373, which were suspected to be in-source fragments of a common precursor. These species were shown to elute in the same chromatographic peak, and switching the instrument polarity from positive to negative ionization mode revealed the common precursor to be cholic acid.
The MarkerLynx XS software package from Waters. A combined scan method aligned the mass spectra, which were then normalized by the constant sum method. Partial least squares-discriminant analysis (PLS-DA) was used to visualize group differences and orthogonal PLS-DA was used to generate S-plots, from which the features contributing the most to the group differences were revealed.
The most challenging aspect was data processing, but not necessarily due to the size of the ESI-IM-MS data set. Ideally, statistical analysis would be performed on data aligned by ion mobility drift times and m/z. However, most conventional software packages rely on peak alignment by chromatographic (liquid or gas) retention times and m/z, and are not yet suited to perform peak alignment by drift times. This is disappointing, as the ion mobility data could not be fully incorporated into the statistical analysis of the wound fluids. As IM-MS becomes more popular, the biostatistics software will likely be developed, which is also an active research pursuit of our group. We anticipate being able to extract even more information from these datasets in the future.
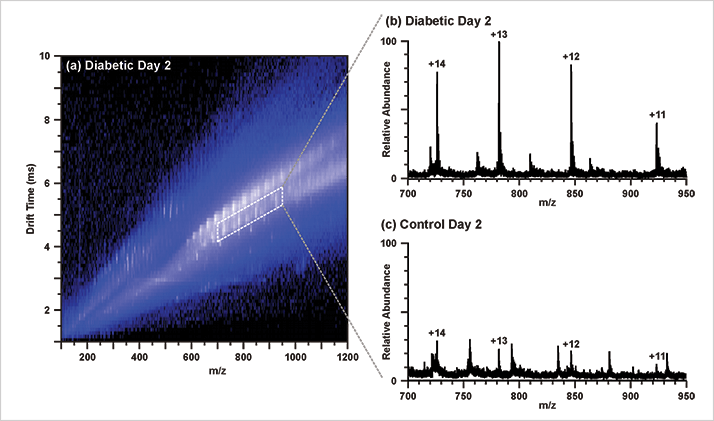
The untargeted approach enables this platform to be used as a discovery tool; tracking treatment studies are one possible avenue to explore. In this particular study, the molecular species differentially expressed between the diabetic and nondiabetic groups, as well as between the diabetic groups at different time points could be useful in this regard.
The IM-MS based approach to identifying biomolecular signatures of disease could be applied to any number of discovery-driven and targeted endeavors due to the unbiased nature of the analysis. In addition to wound fluid, our group has looked at human breast cancer tissue extracts, cerebrospinal fluids, cell lysates, serum, and microbial extracts. RW
Sample Collection and Preparation
Implanted sponges were removed and centrifuged to collect wound fluids. For IM-MS analysis, wound fluids were diluted, desalted, dried and reconstituted in 1:1 methanol: water for electrospray ionization.
System
UHPLC: NanoAcquity UPLC (Waters), Acquity HSS C18 column (1.8 µm, 1.0 x 100 mm). The UPLC analysis was performed online with a Synapt G2
IM-MS (Waters), which utilizes electrodynamic traveling waves and nitrogen gas for ion mobility separations. Mass analysis was performed with orthogonal time-of-flight (oTOF) MS operating in single reflectron mode. The four-dimensional data set comprised of retention time, drift time, m/z and intensity.
References
- K. M. Hines et al., “Biomolecular Signatures of Diabetic Wound Healing by Structural Mass Spectrometry,” Anal. Chem., 85, 3651 (2013) (dx.doi.org/10.1021/ac303594m )




