A breakthrough has been made in the treatment of the fatal skin condition toxic epidermal necrolysis (TEN), made possible through the use of a novel spatial proteomics technique.
In a recent study, researchers in a team led by Matthias Mann (Max Planck Institute of Biochemistry) used deep visual proteomics (DVP) to identify the hyperactivation of the inflammatory JAK/STAT pathway in keratinocytes and immune cells from patients suffering from TEN. This established JAK1 inhibitors as a potential treatment option which, when put in practice, resulted in the rapid re-epithelialization and recovery of affected cells in seven TEN patients.
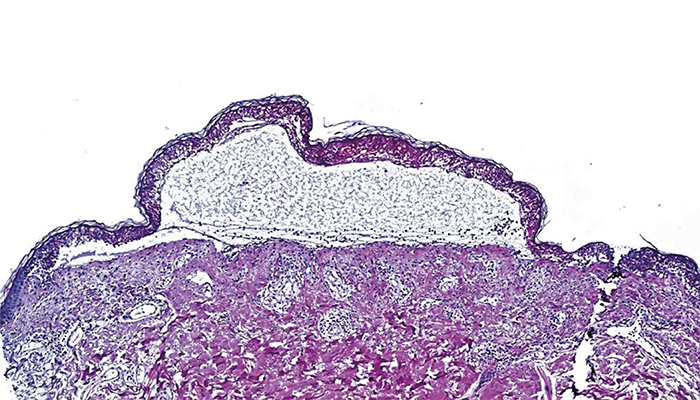
The team applied DVP to formalin-fixed paraffin-embedded (FFPE) skin biopsies from both healthy patients and those suffering from mild to severe skin conditions, such as maculopapular rash (MPR) and TEN. Using AI and ultra-sensitive mass spectrometry, they were able to identify roughly 5,000 unique proteins in each cell type across their participants. From here they could study the disease- specific molecular mechanisms in each cell type under a range of conditions, leading to their eventual discovery.
To find out more, we spoke to first author Thierry Nordmann to discuss the team’s findings, and what he believes they could mean for the future.
Please could you give us an overview of your spatial proteomics approach?
Deep Visual Proteomics (DVP) is a cutting-edge technology that combines microscopy, artificial intelligence, and ultra-sensitive mass spectrometry to analyze specific cell types directly from tissue sections. We applied this approach to study different types of cutaneous adverse drug reactions, ranging from mild maculopapular rash to severe and potentially fatal conditions like toxic epidermal necrolysis (TEN). The beauty of DVP is that it works with formalin-fixed paraffin-embedded (FFPE) tissue, allowing us to use archived diagnostic biopsies to build our patient cohort.
In more detail, we start with a single 3 µm thin section from an FFPE tissue block and perform immunofluorescence staining to label our cells of interest – epidermal keratinocytes and various dermal immune cells including macrophages, CD8+ T cells, and CD4+ T cells. We then scan these stained sections and use a custom-trained artificial intelligence model to identify and classify individual cells. The AI generates precise coordinates for each cell, which we transfer along with reference points to our laser microdissection system. This allows us to return to the exact same locations on the slide and precisely cut out these specific cells. The isolated cells are collected directly into 384-well plates for immediate processing, followed by ultra-sensitive mass spectrometry analysis that identifies and measures thousands of proteins within each cell type. Using this approach, we created detailed molecular maps showing how different cells behave during these severe drug reactions.
What was the biggest challenge you faced during the research?
One of our main challenges was studying such a rare disease as toxic epidermal necrolysis. DVP was a game-changer here because it allowed us to work with FFPE tissue sections, meaning we could build a cohort using retrospective cases from diagnostic biopsies. Laser microdissection of tiny immune cells located in the collagen-rich dermis was the next big challenge, but we managed to do so. Along the way, we even developed new standardized protocols for handling the specialized membrane slides needed for DVP (Nordmann and Schweizer et al., MCP 2023). Another major analytical challenge was achieving reliable mass spectrometry measurements from these microscopically small samples, but conducting this work in the laboratory of Matthias Mann, who developed the DVP technology and is a leader in the field of proteomics, gave us unique insights and capabilities to optimize these sensitive measurements.
Any "eureka moments"?
We actually had three major eureka moments! The first was seeing how beautifully the Deep Visual Proteomics results turned out from these precious and rare cell types, clearly showing the importance of interferon-gamma in immune cells of toxic epidermal necrolysis patients. I was genuinely surprised by just how powerful DVP proved to be – the fact that we could get such detailed molecular insights using archived tissue biopsies was remarkable. Without this capability, we would have needed years to prospectively collect enough samples of such a rare disease. The second eureka moment came when our collaborators in Australia sent us an exciting email showing how well these findings translated to JAK inhibition in mouse models. I literally remember reading the email from Dr. Holly Anderton, a senior scientist at the Walter and Eliza Hall Institute of Medical Research, and thinking, wow, this is major. And of course, the most rewarding moment was seeing how well the patients responded to JAK inhibition treatment, which our collaborators in China implemented.
Can you speak to the potential of spatial proteomics more broadly?
Spatial proteomics represents a powerful new way to understand disease mechanisms at molecular resolution. Our work shows how this technology can make a real difference. We were able to analyze thousands of proteins in specific cell types from archived patient samples, discover a key pathway driving disease, and translate this finding into successful treatment. This direct path from spatial molecular analysis to clinical impact is quite unique in the field of omics technologies. The ability to use archived tissue samples opens up entirely new possibilities – we can now study rare diseases where prospective sample collection would take years, or tackle fundamental questions about cellular aging at molecular resolution. I believe we are at a turning point in translational medicine, and we now have the right tools at the right time to fundamentally change how we understand health and treat diseases.
Image credit: Supplied by Interviewee




