From Ptolemy to Google Maps, humans have been driven to record the landscape around them. By committing our world to paper, we can understand it, order it, and maybe even control it. A blank on a map is intriguing, but unnerving; medieval mapmakers, faced with unknown territories, filled them with ferocious monsters and deadly storms. These mythical beasts were vanquished as intrepid explorers charted the wilderness, filling in the gaps in our knowledge. Can a new kind of cartography help us face down another terror? Cancer is much better understood than it was 50, or even five years ago, but for the millions of people diagnosed with cancer every year – and the doctors who treat them – there are still many troubling uncertainties. Have we caught it in time? Will it spread? What is the best course of treatment? An ambitious five-year project led by the UK’s National Physical Laboratory (NPL) will record the most detailed map yet of the molecular landscape of a tumor. By combining new and existing mass spectrometry imaging techniques, the multidisciplinary team will create a “Google Earth view of cancer” – from whole-tumor down to subcellular level – with the hope of charting a course towards new options for prevention, diagnosis and treatment.
Lead investigator Josephine Bunch shares details of an ambitious project to image all the molecules associated with cancer, in an interview with Charlotte Barker. In 2015, I heard a program on BBC Radio 4 about the Cancer Research UK (CRUK) Grand Challenge – a series of five-year, £20 million awards for multidisciplinary teams willing to take on some of the toughest challenges in cancer research. One of the problems they described was mapping tumors at a molecular and cellular level – creating a Google Earth view of tumors. Traditionally, mapping of tumors was performed using standard histology and histopathology tools, such as microscopy of stained tissues. The problem is that to stain for a molecule, you have to know what it is. To create a comprehensive map, we have to be able to find all molecules, including the unexpected. As an analytical scientist, I saw that what CRUK was describing was fundamentally a measurement challenge, and felt immediately that the way forward would be through mass spectrometry imaging (MSI). Admittedly, I may have been somewhat biased. From the moment I first encountered mass spectrometry, I was hooked. I love the sheer variety of instruments available; the many ways we can create and transmit ions gives us a huge number of different combinations. Then there’s the breadth of applications – mass spectrometry measurements are being recorded everywhere from oceans to operating theaters to missions on Mars. At the time, I had been Co-Director of the National Physical Laboratory (NPL)’s National Centre of Excellence in Mass Spectrometry Imaging (NiCE-MSI) for three years, leading our efforts in ambient MSI and matrix assisted laser desorption/ionization (MALDI). Friends and colleagues encouraged me to contact CRUK to see if they would consider a MSI-based project. CRUK confirmed that there were no preconceived ideas about how the challenge should be solved, and I decided to go for it.
Google Translate
First, I put together a consortium of researchers, some of whom I already knew or had heard of, and others who were recommended to me. The team consists of experts in the relevant cancers, world-leaders in developing genetic models, inventors and innovators of techniques, and specialists in various aspects of tumor biology and metabolism. We will take samples from breast, pancreatic and colorectal cancer, from patient biopsies and mouse models, and we will use them to build chemical images – molecular maps – at a range of different scales, from single cells up to whole tumors. The reason we often use CRUK’s clever analogy of “Google Earth for tumors” is because it is so important to be able to explain our work in accessible terms; partly to communicate the value of our research to the public, but also so that our team of analytical chemists, physicists, biologists and medics are able to articulate shared goals. We all want to achieve the same thing but we don’t always speak the same language. For example, if you ask a biologist about the biggest challenge in mapping a tumor, they are likely to mention the difficulties of obtaining samples for molecular or metabolic studies, and interpreting the information. A mass spectrometrist may have a completely different answer, focusing on the huge sample numbers involved or the problem of building instruments with the resolution required. The Google Earth analogy has guided us in designing our pipeline and bringing the right investigators and techniques on board. I believe that the better you can break the project down into accessible descriptions across disciplines, the better the science.A grand measurement challenge
Getting the call to say we’d been successful in our bid was incredibly exciting. NPL is leading the consortium and will be analyzing samples with MALDI, desorption electrospray ionization (DESI), secondary ion MS (SIMS), Nano-SIMS and OrbiSIMS. We will also be coordinating imaging performed at other institutions, managing the data generated and disseminating the resulting protocols and instrumentation. We launched the project officially in May 2017, and even before that we were getting our instruments ready and gathering preliminary data. Our results so far have shown the extraordinary amount of data possible when we combine different MSI techniques. However, there is also an abundance of challenges, from ensuring that we have sensitivity at the highest resolutions for key metabolites to maintaining the quality of each and every measurement. But mining the enormous data sets we collect is perhaps the biggest challenge of all. Our priority for now is to build a framework to ensure quality measurements across the huge number of samples we plan to analyze. From sample collection to data analysis, there are so many factors that could introduce variation, especially when working with multiple techniques. NPL and the Grand Challenge consortium are extremely passionate about generating reproducible data and repeatable measurements. We don’t want to produce beautiful pictures that cannot be reproduced or don’t accurately represent the underlying cancer biology.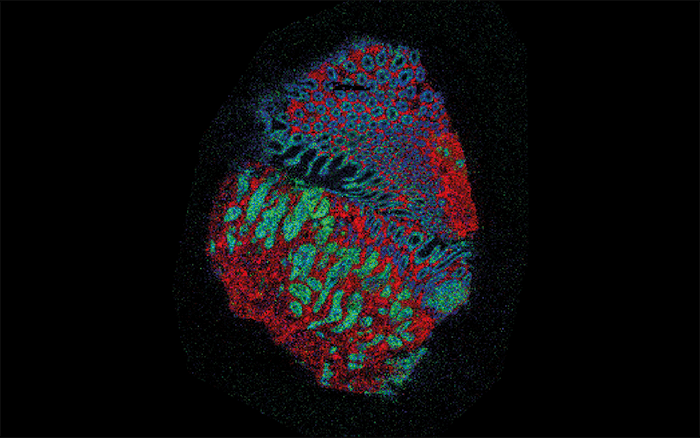
We have designed pilot studies assessing the performance of the plethora of different MSI instruments we’re using – and we’ve made some measurements at several different sites to understand how much it affects the results. This work represents an important foundation in being able to quote the performance of the different techniques in combination. We have also been assessing the various parts of our pipeline to ensure absolute consistency. It is perhaps not the most glamorous work, but it’s vital that samples from different sites are being collected, stored, transported and analyzed in the same way, and that we have robust pipelines in place for handling our data all the way from raw files to the curated data that will be available to the public. Right now, I’m most excited about making measurements that no-one has ever made before whilst working with an extraordinary consortium of researchers – all of whom are there to ensure that we are making measurements that matter. I’m also looking forward to the second phase of the project: interpreting our data, sharing it and broadening our network. Over the five years, we hope to share results so exciting that other labs are inspired to use our techniques. And we will be ready to help them acquire quality data as quickly as possible.
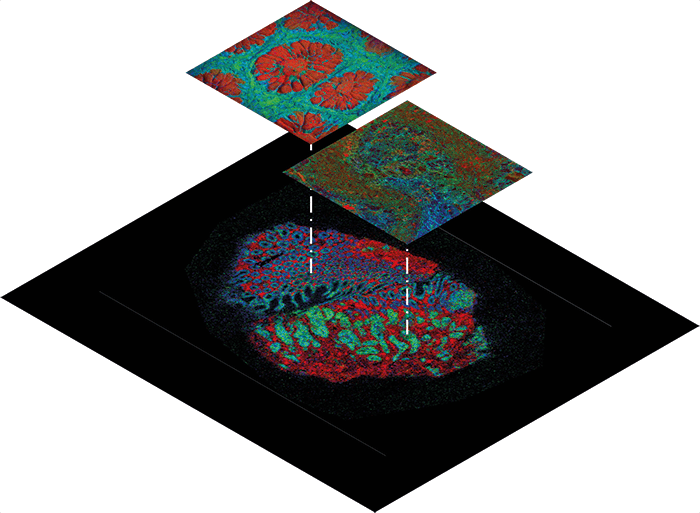
Next-level MSI
The past few years have seen fantastic work from around the world using MSI as a powerful method for mapping multiple molecules in the same tissue. It might still be an emerging technique, but it already has a fantastic pedigree of excellent results. The Grand Challenge project will build on that success by using several MSI techniques in combination to widen the range of molecules examined, and so gain an in-depth understanding of tumor metabolism – linking genes, proteins, peptides, lipids and metabolites. We will be making use of significant recent advances in technology. An example is the 3D OrbiSIMS instrument, which combines SIMS with an Orbitrap mass analyzer. As Ian Gilmore describes in “The Super-Resolution Revolution,” the hybrid instrument allows very high resolution. Going back to the Google Earth analogy, OrbiSIMS is like peering through the window of a house in Street View to see where the sofa is... At that level of detail you can’t get through enormous numbers of samples – just as you wouldn’t want to record the position of every sofa in the world. Instead, we will use other techniques to identify cells and regions within the tumor that we want to pay special attention to. MALDI and DESI give a street and city view – they can get down to pixel sizes small enough for a street-level look but can also rapidly create a basic “city map” of a tumor section. Other techniques will help us localize our search to specific “cities”. In addition to MSI methods, such as SIMS, MALDI and DESI, we are also using techniques for in vivo analysis and imaging of metabolites – such as the iKnife (REIMS) and MRI. A common aspect of all the techniques is that they will produce a series of mass spectra acquired at discrete locations across the tissue. We are developing methods to handle this enormous hyperspectral data set, from raw files to basic pre-processing, such as peak alignment and peak picking, to reduce the volume of data. We may also need strategies for normalization so that signals collected on different days can be meaningfully compared. The real challenge comes when we want to mine those data; we will need to use a whole range of machine-learning tools, linear and non-linear methods to group similar samples together, to segment areas of relevance, and to try to understand associations between the molecules detected.Measuring success
Our ultimate goal is to gain new insights into tumor progression that might help diagnose and treat cancer. If we can help biologists understand exactly how tumors grow and spread, that knowledge can be translated to make sure that patients are diagnosed earlier, and can be given the right treatments at the right time. Of course, it will take time to translate our findings into the clinic. Concentrating on the next five years, I will be satisfied if:- The tumor biologists on the team have gained fresh understanding.
- We have significantly improved the performance of the techniques we’re using.
- Our measurements are being adopted as standard in research labs.
- Our data have helped to produce new in vitro models that are more representative of real human tumors.
A diverse group of analytical chemists, physicists, biologists and medics have come together to make the vision of a Google Earth for tumors a reality. Josephine Bunch Bunch will lead the Grand Challenge consortium. She is Co-Director of the National Centre of Excellence in Mass Spectrometry Imaging (NiCE-MSI) at NPL. Ian Gilmore Gilmore is a Senior NPL Fellow and Head of Science at NPL. He is the founder of the National Centre of Excellence in Mass Spectrometry Imaging (NiCE-MSI) at NPL, where he conceived of the 3D OrbiSIMS instrument and led the project to build it. The Gilmore group will lead the high-resolution 2D and 3D SIMS imaging for the Grand Challenge. John Marshall A professor of tumor biology at Barts Cancer Institute, Queen Mary University in London, Marshall is an expert in tumor invasion and the role of adhesion molecules. In the Grand Challenge, the Marshall group will deliver imaging CyTOF (mass cytometry) analysis. Owen Sansom Sansom is interim director of the Cancer Research UK Beatson Institute. He has been instrumental in determining the molecular hallmarks and cell of origin of epithelial cancers (colorectal and pancreatic). The Sansom laboratory will provide the Grand Challenge team with in vivo models. Richard Goodwin A Principal Scientist at AstraZeneca, Goodwin leads a MSI group studying the distribution of drugs. Within the Grand Challenge team, his role is to help perform inter-site experiments, translate the findings into an industry setting and disseminate for maximum impact on the development of new oncology medicines. Mariia Yuneva Yuneva leads a group at the Francis Crick Institute dedicated to oncogenes and tumor metabolism. In the Grand Challenge, her group will provide in vivo and ex vivo models of mouse and human primary breast cancers and their metastases. George Poulogiannis Poulogiannis joined the Institute of Cancer Research (ICR) in 2014 and now leads the Signaling and Cancer Metabolism team. His contribution to the Grand Challenge will be to study the therapy sensitivity pattern of metabolically-distinct tumor phenotypes using genetic and pharmacological approaches. Zoltan Takats The inventor of multiple analytical methods for direct analysis of biomolecular systems, including the iKnife, Takats is Professor of Analytical Chemistry at Imperial College London. In the Grand Challenge, the Takats group will help deliver multi-modal MSI, and lead REIMS and iKnife studies. Kevin Brindle Brindle is Professor of Biomedical Magnetic Resonance at the University of Cambridge and a senior group leader in the CRUK Cambridge Institute. In the Grand Challenge, the Brindle group will be responsible for hyperpolarized 13C imaging in the clinic, collection of tumor material in surgery and production of patient-derived orthotopic tumor xenografts. Simon Barry Barry is a Senior Principal Scientist in the IMED Oncology group at AstraZeneca. His research focuses on the cross talk between the tumor and its micro-environment. AstraZeneca’s Oncology group will support the Grand Challenge with specialist technical and scientific contributions. Kelly Gleason For the last 12 years, Gleason has led a team of research nurses in the field of oncology clinical research. She has also supported the Imperial Patient and Public Involvement (PPI) Group for the Imperial CRUK Centre for the past five years. Harry C. Hall After a colorectal cancer diagnosis in 2002, Hall became a founder member and chair of W London Cancer Network Partnership Group. He now sits on the NIHR Imperial BRC PPI Panel, Imperial College & Partners PPI Research Forum and the CRUK Imperial Centre PPI Group.
Josephine Bunch Bunch will lead the Grand Challenge consortium. She is Co-Director of the National Centre of Excellence in Mass Spectrometry Imaging (NiCE-MSI) at NPL.

Ian Gilmore Gilmore is a Senior NPL Fellow and Head of Science at NPL. He is the founder of the National Centre of Excellence in Mass Spectrometry Imaging (NiCE-MSI) at NPL, where he conceived of the 3D OrbiSIMS instrument and led the project to build it. The Gilmore group will lead the high-resolution 2D and 3D SIMS imaging for the Grand Challenge.

John Marshall A professor of tumor biology at Barts Cancer Institute, Queen Mary University in London, Marshall is an expert in tumor invasion and the role of adhesion molecules. In the Grand Challenge, the Marshall group will deliver imaging CyTOF (mass cytometry) analysis.

Owen Sansom Sansom is interim director of the Cancer Research UK Beatson Institute. He has been instrumental in determining the molecular hallmarks and cell of origin of epithelial cancers (colorectal and pancreatic). The Sansom laboratory will provide the Grand Challenge team with in vivo models.

Richard Goodwin A Principal Scientist at AstraZeneca, Goodwin leads a MSI group studying the distribution of drugs. Within the Grand Challenge team, his role is to help perform inter-site experiments, translate the findings into an industry setting and disseminate for maximum impact on the development of new oncology medicines.

Mariia Yuneva Yuneva leads a group at the Francis Crick Institute dedicated to oncogenes and tumor metabolism. In the Grand Challenge, her group will provide in vivo and ex vivo models of mouse and human primary breast cancers and their metastases.

George Poulogiannis Poulogiannis joined the Institute of Cancer Research (ICR) in 2014 and now leads the Signaling and Cancer Metabolism team. His contribution to the Grand Challenge will be to study the therapy sensitivity pattern of metabolically-distinct tumor phenotypes using genetic and pharmacological approaches.

Zoltan Takats The inventor of multiple analytical methods for direct analysis of biomolecular systems, including the iKnife, Takats is Professor of Analytical Chemistry at Imperial College London. In the Grand Challenge, the Takats group will help deliver multi-modal MSI, and lead REIMS and iKnife studies.

Kevin Brindle Brindle is Professor of Biomedical Magnetic Resonance at the University of Cambridge and a senior group leader in the CRUK Cambridge Institute. In the Grand Challenge, the Brindle group will be responsible for hyperpolarized 13C imaging in the clinic, collection of tumor material in surgery and production of patient-derived orthotopic tumor xenografts.

Simon Barry Barry is a Senior Principal Scientist in the IMED Oncology group at AstraZeneca. His research focuses on the cross talk between the tumor and its micro-environment. AstraZeneca’s Oncology group will support the Grand Challenge with specialist technical and scientific contributions.

Kelly Gleason For the last 12 years, Gleason has led a team of research nurses in the field of oncology clinical research. She has also supported the Imperial Patient and Public Involvement (PPI) Group for the Imperial CRUK Centre for the past five years.

Harry C. Hall After a colorectal cancer diagnosis in 2002, Hall became a founder member and chair of W London Cancer Network Partnership Group. He now sits on the NIHR Imperial BRC PPI Panel, Imperial College & Partners PPI Research Forum and the CRUK Imperial Centre PPI Group.

Josephine Bunch Bunch will lead the Grand Challenge consortium. She is Co-Director of the National Centre of Excellence in Mass Spectrometry Imaging (NiCE-MSI) at NPL.





