Food fraud is not going away – a fact that was cemented by the horsemeat scandal that first surfaced in early 2013. How can speciation testing be improved to quell consumer fears about the authenticity of meat?
At the height of the horsemeat scandal, supermarket chains were forced to announce that some “beef” products actually contained up to 100% horsemeat. This led to concerns about the regulations that were in place for products entering the human food chain and cast serious doubt on the ideal of “farm to fork”; many people stopped trusting what they had been happily eating, resulting in calls from governments, industry and, of course, consumers for a better way to guarantee the authenticity of meat.
The substitution of beef products with horsemeat is likely to have been motivated by cost – what else? Pork, which also trades at a lower price than beef, has also been fraudulently substituted (possibly along with more unsavory species…). And while contamination of beef with meat from other species is unappealing to many people, it can be especially distressing for those with strict religious beliefs. Judaism and Islam both forbid eating food containing any type of pig meat. It follows that Jewish and Muslim communities need to know of any contamination of foods that are otherwise considered permissible to eat (kosher or halal, respectively). Beyond ethical or religious grounds, microbiological and chemical product safety is also at risk with such blatant disregard of food regulations. We need to authenticate our meat. Common methods that are widely used for meat authentication testing include polymerase chain reaction (PCR) and protein assays, such as enzyme-linked immunosorbent assays (ELISA). These techniques are fairly quick and reproducible, but not always reliable. PCR is one of the most commonly used methods for meat speciation testing. And though it generally allows for the detection of different species of meat simultaneously, the quality of the sample is important; any sample DNA degradation can be problematic. Unfortunately, such degradation can occur during certain meat processing and food manufacturing processes, giving plenty of potential for contaminants to remain undetected.
Protein assays are also commonly used because they are widely available and easy to use. However, it is difficult analyze for more than one protein marker at a time, making the approach time-consuming and somewhat limited. ELISAs require a greater sample volume because of the difficulties in multiplexing assays. And the generation of false positive or false negative results (caused by cross-reactivity of antibodies or by sample processing, respectively) is a final big issue. PCR and ELISA sensitivity is considered “acceptable” – that is to say, it meets the the UK’s Food Standards Agency (FSA) guidelines , for example, which recommends a one percent threshold for reporting cross-species contamination of meat. Some in the Jewish and Muslim communities would say that the requirement is to provide quantitative results at the lowest limit of detection possible – a view shared by others with a zero-tolerance policy towards meat contamination.
Scientists at the University of Münster, Germany, and AB Sciex in the UK have developed an alternative speciation method for detecting pig, horse or beef proteins in meat. The method was published recently in the Journal of Agriculture and Food Chemistry (1).
Hardware: Tandem Mass Spectrometry (MS/MS) was carried out using the QTRAP 5500 LC-MS/MS system (AB SCIEX)
Software: The iterative workflow of the AB SCIEX MRMPilot software, version 2.1 was used to develop the MRM method.
The possible MRM transitions were identified and introduced into the initial method using predicted MS/MS spectra of target peptides. Purified peptide extracts from respective species or synthesized peptides were used to determine the retention time of target peptides and optimization of collision energy (CE). The most intense transitions were then identified and optimized for a second time.
To enhance sensitivity further, a micro-LC system was used. Positive control samples from respective species were used to assess the retention time stability and relative intensity of MRM transitions.
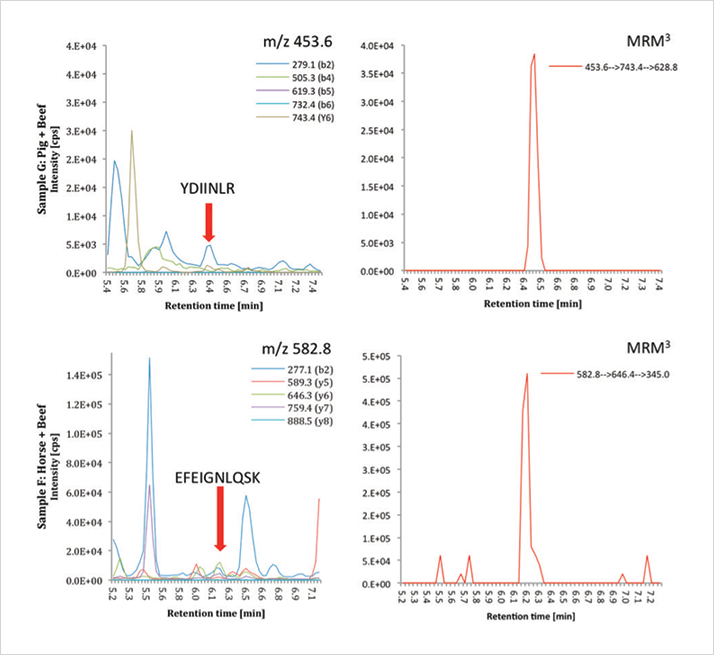
A key objective of the work was to develop a solution that is easy to use. The new method, which uses liquid chromatography (LC) and tandem MS (LC-MS/MS), does exactly that while also offering a more accurate, reliable approach to meat speciation than other methods. Steve Lock, Business Development Manager at AB Sciex, suggested in 2012 that: “Mass spectrometry could potentially be used to look for over 10 different species in one go, as has previously been shown in the detection of 16 allergenic species in one analyses” (2). Indeed, the potential for detecting multiple, different animal species in a single run is a further advantage over currently accepted methods. The first step in method development was to identify species-specific polymorphisms in proteins that would be detectable by MS. The research group at the University of Münster focused on myofibrillar and sarcoplasmic proteins because they are highly abundant and, therefore, allow higher sensitivity. The team extracted protein fractions from commercially available meat samples, including cow, pig, wild boar, horse, chicken and lamb, and used high-resolution MS (HRMS) to identify species-specific biomarker peptides. The second step was to confirm the presence of targeted meat peptides in unknown samples. A multiple reaction monitoring (MRM) approach was taken, which is capable of providing sequence information, allowing peptides to be identified (see “The System”). The proteins found in horse and beef meat, for example, may differ by only two amino acids; mass spectrometry can detect these differences and indicate where they are in the sequence, avoiding the risk of false positives. The most abundant biomarker peptides for horse, beef, and pork identified by the non-targeted proteomic approach were used to develop the MRM method. The MS parameters and conditions were optimized, and species specificity confirmed for all chosen peptides. The three most intense marker peptides identified are shown in Table 1. These abundant biomarkers can be used for the detection of trace contaminations of pork or horse in beef.
Using MRM, the detection limit of horse spiked into beef was 0.55 percent. To achieve higher sensitivity and enhance the signal-noise ratio, the MRM3 mode of the QTRAP system was used. This allowed detection of pig and horse down to concentrations of less than 0.25 percent. Figure 1 shows the detection of two peptide sequences in beef using MRM3.
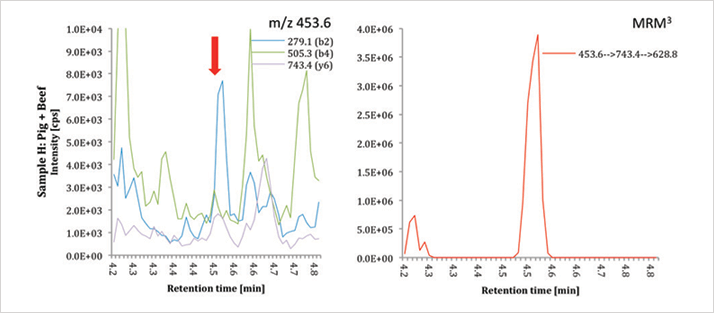
The sensitivity of the method was enhanced even further, by using the QTRAP 6500 system (equipped with micro-LC) to detect 0.13 percent pork in beef (see Figure 2).
In total, the scientists at the University of Münster identified 12 tryptic biomarker peptides specific for pork and/or horse meat. To their knowledge, they were the first to use MRM or MRM3 as a sensitive and rapid MS-based technique. The sensitivity of the method is comparable with the most sensitive PCR and ELISA methods and uses only a small amount of sample. Another advantage of the approach is that it does not require extensive pre-fractionation for proteome characterization.
The new method can be used to test meat products quickly and easily for the presence of pork, horse and beef.
Next steps include identifying peptide biomarkers for other species, and the subsequent development of a multi-species identification tool. The method will be optimized for foods that have been heavily processed (for example, pre-cooked); unprocessed (raw) meat was used in experiments outlined here and the sensitivity of the method is likely to be reduced in processed samples or low-quality meat.
At present, MS is often over looked. According to Hans-Ulrich Humpf of the University of Münster, “Many labs have the QTRAP system, but customers don’t always realize the full power and capabilities of the QTRAP”. But analysis is evolving and, given its many advantages, it is likely that MS-based techniques will be increasingly common in food analysis labs. In fact, MS techniques are already being used for allergen testing in food and wine.
Another future development will be the detection of gelatin from cows, pigs or horses in non-meat foods, which will be of particular interest to those with specific dietary preferences. Watch this space.
References
- C. von Bargen et al., “New Sensitive High-Performance Liquid Chromatography–Tandem Mass Spectrometry Method for the Detection of Horse and Pork in Halal Beef”, J. Agric. Food Chem., 61 (49), 11986–11994 (2013). S. Lock, “Allergen screening in Food by LC/MS/MS”, conference proceedings 126th AOAC Annual meeting, Las Vegas 2012.




