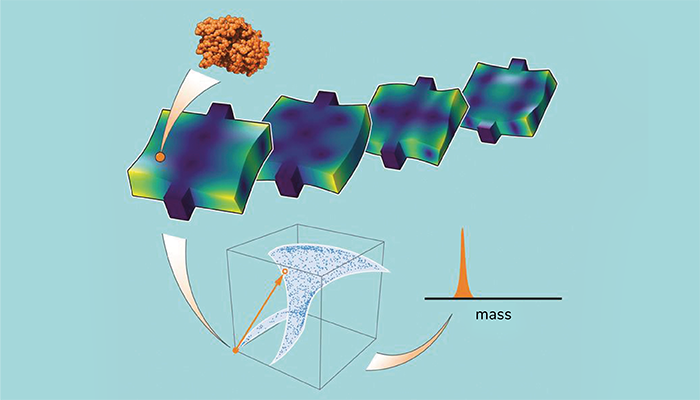
In Caltech's new fingerprint technique, a single molecule adsorbs onto the phononic crystal resonator device. Then scientists measure the frequency shifts of four different vibrational modes of the device, allowing them to create a four-dimensional fingerprint vector—a unique identifier that can then be used to determine the mass of the molecule.
Credit: Nunn/Caltech
Nanoelectromechanical systems (NEMS) can directly measure the mass of an analyte without the need for prior fragmentation or ionization – a significant advantage over standard mass spec methods. NEMS, however, often needs prior knowledge of an analyte’s mode shapes to produce accurate results, which isn’t always attainable.
To navigate this issue, researchers from the California Institute of Technology have developed a machine learning-based approach, referred to as fingerprint nanoelectromechanical mass spectrometry, which allows them to accurately measure the mass of proteins at a molecular level through the use of nanoscale devices.
Their main motivation? To repeat the success of the Human Genome Project for proteins. We spoke to co-authors John Sader and Michael Roukes to find out more.
Can you explain, in a nutshell, how your technique works?
The mass of a biomolecule or particle can be measured by placing it on a vibrating NEMS device and measuring the change in frequency of the device. A heavier device will vibrate at a lower frequency, for example. This conventional approach requires the shape of several vibrating modes to be known. This isn’t always possible however, especially for the latest NEMS devices that promise extreme mass resolution.
Our technique dispenses with this (standard) mode shape requirement using a machine learning approach. First, a particle of known mass is placed on the NEMS device at different positions to build up a library containing the frequencies of its many vibrational modes. The mass of an uncharacterized particle can then be measured by comparing the vibrational frequencies that it induces on the same NEMS device to those within the library. This allows the use of any device to measure a particle’s mass, as it does not require the mode shapes.
Was there a “eureka moment” during development?
Yes – our theory team had been brainstorming how to measure the mass of biomolecules using NEMS devices with unknown vibrational modes. We realized that this was the major obstacle to using the latest advanced devices with ultrahigh mass sensitivity. Then during one discussion, a left field mathematical idea popped up: to construct an array of numbers (a vector) from the measured frequency shifts of different vibrational modes of the NEMS device. This “fingerprint” vector was hypothesized to depend uniquely on where the particle landed – later we mathematically proved this to be true! This was the foundation of the new fingerprint approach, and was validated in the experiments we reported in our paper.
Can you speak to the ultimate potential of this technique? For example, there are initiatives, such as the Human Proteoform Project, that are aiming to repeat the success of the Human Genome Project for proteins; could your technique play a role here?
Absolutely. This is directly in our sights and a principal motivation for our work. Our aim is to enable deep profiling of the single-cell and serum proteomes, as well as the exploration of protein-protein interactions. The latter of these is generally inaccessible by bottom-up mass spectrometry, which disassociates proteins into peptide fragments to enable analysis. With NEMS aided by this machine learning method, we can advance the technology for single-molecule native mass spectrometry – that is, mass spectrometry on intact large proteins and protein complexes which preserves these delicate interactions.
Are there any major hurdles you’ll have to overcome to realize this potential?
We don’t foresee any at present! Our next steps are to implement the new method into our native, single-molecule mass spectrometry workflow. Currently, we are doing just that!
What are your next steps for this work?
We are currently working with our colleague, Prof. Amir Safavi-Naeni from Stanford University, to apply the fingerprint approach to his phononic crystal NEMS devices.
We’re excited to see our new technique in action which will allow mass measurements on these advanced NEMS devices for the first time.




