
Chemical interactions can drive cell function. Because of the striking chemical heterogeneity found within cell populations, analyzing cells individually can uncover mechanisms not observable when studying the chemistry from homogenized cellular populations. However, the intricacies of single cell investigation become more overwhelming the more we consider them; from sample preparation to analysis, smaller scales increase our risk of failure.
For decades, the hyphenation of approaches, often incorporating volume-matched separations, has aided chemical measurements (1). So it’s not surprising that methods to analyze individual cells using multiple combined measurement techniques have expanded our capabilities. Here, we highlight several serial approaches that boost the information obtained from single cell analyses.
Separating cellular complexities
Our lab has spent many years creating measurement technologies using capillary electrophoresis (CE) to explore the chemistry of volume-limited samples – in some cases, even smaller than single cells (2). We have characterized single neurons and subcellular features, and uncovered chemical complexity in animal models ranging from mollusks to mammals. CE requires nanoliter volumes and thus reduces sample dilutions from single cell separations (3). By hyphenating CE to mass spectrometry (MS) (4), we have expanded our ability to explore complex single cell chemistry and characterize both familiar and new compounds from selected cells.
The traditional and easiest methods for CE sample preparation involve placing the sample into an extraction buffer or, in some cases, injecting the entire cell into the capillary. The limited sample remaining after a measurement often precludes follow-up analysis, especially once technical replicates are performed. Accordingly, we and other scientists have gotten creative in our efforts to develop sampling techniques that reduce analyte losses and enable multiple distinct measurements. For example, we used patch-clamp electrophysiology to identify and characterize neurons, and then used the same patch-clamp pipette to extract a few picoliters of cell cytoplasm for follow-up CE-electrospray ionization (ESI)-MS metabolite profiling (5). Other labs have used similar micropipette sampling techniques, leaving much of the cell intact and alive (6).
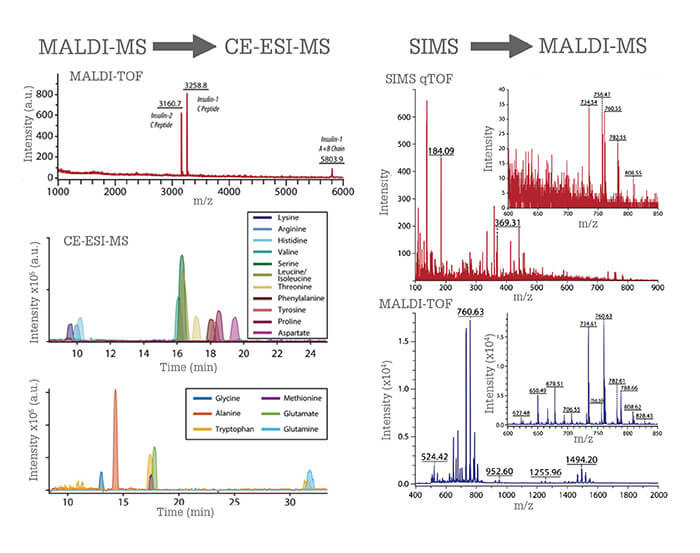
More recently, we developed a liquid microjunction (LMJ) sampling probe that enables the extraction of cellular content from cells located on a microscope slide (7). The probe consists of two concentric capillaries; solution is pumped through the outer capillary and aspirated through the inner capillary. Cellular material is collected as fluid migrates from the outer to inner capillary. Analysis can be carried out before or after metabolite extraction, depending on our needs and the measurement technique being used. Not only can we perform CE-ESI-MS metabolite profiling, we can add other minimally destructive slide-based chemical measurements of the same cell. We validated the approach by hyphenating matrix-assisted laser desorption/ionization (MALDI) MS to CE-ESI-MS to analyze single cells from rat pancreatic islets of Langerhans, micro-organs that perform the canonical glucose-regulating functions of the pancreas. Pancreatic cell types are defined by the presence of a peptide hormone; beta cells contain insulin and alpha cells contain glucagon. We screened cells for peptides (revealing cell type) using MALDI MS, selected the cells of interest, and extracted the small molecule metabolite content with the probe. We then performed follow-up metabolite profiling with CE-ESI-MS, reporting one of the first direct detections of canonical neurotransmitters in single pancreatic alpha and beta cells. Our ability to perform these combined measurements has opened the door to studying pancreatic chemistry from human islets used in islet transplantations.
Sample savior
The crux of serial analyses for single cell studies lies in the preservation of cellular material from the first analysis for use in the next measurement. An early study in our lab established that at least 60 percent of cellular material remains on the surface following MALDI MS analysis (8). Although the laser shots do consume cellular contents, the extent is less than most think, and MALDI MS measurements do not preclude the cell from being re-assayed.
To achieve a more robust reanalysis, we created microMS (9), open-source cell-finding software that enables single cell targeting on a microscope slide. Multiple research groups have developed strategies to target single cells beyond simply imaging the entire slide. In our approach, we use optical imaging to register the spatial locations of cell nuclei on a slide and then direct the laser, electron beam, or LMJ probe to the desired cell locations. During development of the software we also determined that, in addition to leaving chemical material behind, such analyses do not displace the cell.
The microMS software enables single cell analysis on a high-throughput scale; we can analyze thousands of cells in several hours. We have performed sequential profiling using several different mass spectrometers: MALDI time-of-flight (TOF), MALDI Fourier transform-ion cyclotron resonance (FT-ICR), and C60+-secondary ion mass spectrometry (SIMS). We can leverage the advantages of each instrument to collect complimentary information for single cell samples; for example, lipid and peptides via MALDI-TOF MS, high-resolution spectra and elemental composition confirmation with MALDI-FT-ICR MS, and small molecule content from SIMS (Figure 1).
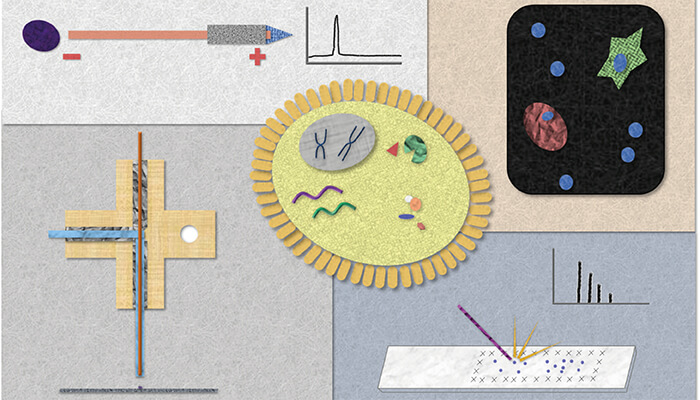
A wider world of measurements
Our lab has employed various strategies to hyphenate MALDI MS to a number of analytical approaches, including immunocytochemistry, spectroscopy, and transcriptomics. Despite the potential for sample destruction, by carefully sequencing our experiments we can add the advantages inherent to each individual technique.
For instance, immunostaining for proteins is the gold standard method for cell classification, including defined cell types such as neurons, astrocytes, and other brain cells. However, immunostaining requires fixation of the cells, which renders much of the cellular chemistry difficult to characterize because of the crosslinking of proteins, peptides, and even lipids and sugars. This presents a challenge in validating our single cell separations and MS data.
We address this issue by performing single cell MALDI MS prior to fixation and immunostaining. In addition, in collaboration with the Bhargava laboratory, we have combined vibrational spectroscopy with both mass spectrometry imaging (10) and single cell measurements to provide enhanced information on lipids, nucleotides, and proteins.
In summary, we have enhanced our ability to investigate a population of cells using more than one omics measurement. We can sequentially measure an individual cell using multiple distinct approaches. By analyzing a single cell multiple times using techniques suited for distinct classes of molecules, we further our ability to probe biological systems on the cellular scale. Combining these measurements dramatically increases the potential for advancement while creating an analytical toolkit that is as diverse as the chemistry within a single cell itself.
By Kevin Schug, Conference Chair
This exciting work by Philip and Sweedler is a perfect example of the cutting-edge research and technical advancements on display at the 43rd International Symposium on Capillary Chromatography and the 16th GCxGC Symposia held in Fort Worth, Texas, in May. Professor Sweedler will be one of several high-profile plenary speakers amongst a vibrant and well-attended scientific program. Analytical science is moving to smaller scales, and to multidimensional and automated platforms, trends that will be front and center at the event. Here, you can both learn the fundamentals from the world’s leading experts and view the newest advances in technology and applications. Not too big and not too small, ISCC & GCxGC 2019 provides ample opportunity to rub shoulders with key opinion leaders in a relaxed and welcoming atmosphere, underscored by a Texas-style social program. Register now to attend the key event of 2019 for advances in capillary and comprehensive separation technologies.
Plus, abstract submission for both oral and poster sessions is currently open – apply now to join the conversation!
The 43rd International Symposium on Capillary Chromatography (ISCC) and the 16th GCxGC will be held in Fort Worth, Texas USA on May 13, 2019. Register at www.isccgcxgc.com
References
- T Hirschfeld, “The hy-phen-ated methods”, Anal Chem, 52, 297A–312A (1980). https://doi.org/10.1021/ac50052a870. SS Rubakhin et al., “Measuring the peptides in individual organelles with mass spectrometry”, Nat Biotechnol, 18, 172–175 (2000). https://doi.org/10.1038/72622. JA Jankowski et al., “Assaying single cells with capillary electrophoresis”, Trends Anal Chem, 14, 170–176 (1995). https://doi.org/10.1016/0165-9936(95)98315-Y. T Lapainis et al., “Capillary electrophoresis with electrospray ionization mass spectrometric detection for single-cell metabolomics”, Anal Chem, 81, 5858–5864 (2009). https://doi.org/10.1021/ac900936g. JT Aerts et al., “Patch clamp electrophysiology and capillary electrophoresis–mass spectrometry metabolomics for single cell characterization”, Anal Chem, 86, 3203–3208 (2014). https://doi.org/10.1021/ac500168d. RM Onjiko et al., “In situ microprobe single-cell capillary electrophoresis mass spectrometry: metabolic reorganization in single differentiating cells in the live vertebrate (Xenopus Laevis) embryo”, Anal Chem, 89, 7069–7076 (2017). https://doi.org/10.1021/acs.analchem.7b00880. TJ Comi et al., “MALDI MS guided liquid microjunction extraction for capillary electrophoresis–electrospray ionization MS analysis of single pancreatic islet cells”, Anal Chem, 89, 7765–7772 (2017). https://doi.org/10.1021/acs.analchem.7b01782. JS Page, JV Sweedler, “Sample depletion of the matrix-assisted laser desorption process monitored using radionuclide detection”, Anal Chem, 74, 6200–6204 (2002). https://doi.org/10.1021/ac025898k. TJ Comi et al., “MicroMS: a Python platform for image-guided mass spectrometry profiling”, J Am Soc Mass Spectrom, 28, 1919–1928 (2017). https://doi.org/10.1007/s13361-017-1704-1. EK Neumann et al., “Multimodal chemical analysis of the brain by high mass resolution mass spectrometry and infrared spectroscopic imaging”, Anal Chem, 90, 11572–11580 (2018). https://doi.org/10.1021/acs.analchem.8b02913.




