We Need More Structure
Protein analysis using non-destructive NMR can elucidate how proteins interact with drugs and help in the development of pharmaceutical treatments
Tatyana Polenova | 8 min read | Discussion

Delving into the world of protein structure, dynamics, and interactions offers a groundbreaking pathway to understanding disease mechanisms. Comprehensive knowledge of these intricate biomolecules, particularly related to disease, holds the potential to revolutionize targeted therapy and drug development. By uncovering the precise roles and dysfunctions of specific proteins, normal cellular function can be restored, paving the way for therapeutic advancements.
The pharmaceutical landscape faces significant challenges. A study conducted from 2001 to 2020 revealed that during those two decades pharmaceutical companies increased their research and development spending by six percent annually, launched 251 drugs and spent $6.16 billion per drug approval (1). The increased expenditure of bringing drugs to market outlines the imperative to optimize development timelines and reduce costs.
Emerging technologies are addressing these concerns, providing researchers with cutting-edge tools to elucidate the intricacies of the protein landscape. Since the action mechanism of nearly every drug involves binding to a protein, gathering information about these proteins facilitates drug research. In this respect, techniques like nuclear magnetic resonance (NMR) offer unparalleled structural insights, capturing proteins in their native states and providing intrinsic detail. By observing their dynamic movements and interactions, valuable knowledge is gained about their functional roles and dysfunctions in disease.
Exploring protein dynamics with advanced technologies
Deciphering the three-dimensional shapes and dynamic behaviors of proteins allows researchers to identify structural anomalies or alterations specific to disease-related proteins, providing insights into the molecular foundations of various disorders. Understanding these conformational fluctuations and their impact on protein–protein interactions presents a significant challenge, demanding a multifaceted approach using various biophysical techniques (2).
Multiple biophysical techniques, each with their own strengths and limitations, are employed to analyze and determine protein structures. X-ray crystallography, a popular method for revealing the three-dimensional atomic structure of proteins, analyzes the diffraction patterns of X-rays passed through a crystal lattice. But because of the inherently dynamic nature of proteins, it can be challenging to capture all of these structural states using this method alone. Additionally, obtaining high-quality protein crystals suitable for X-ray crystallography can be time consuming and, in some cases, impossible.
Cryo-electron microscopy (cryo-EM) has emerged as another useful tool for protein structure determination. This technique visualizes biological macromolecules and structures at near-atomic resolution by freezing samples in vitreous ice and capturing detailed images using electron microscopy (3). Despite its high-resolution capabilities, cryo-EM faces challenges when dealing with intrinsically disordered proteins or those lacking well-defined three-dimensional structures, making it difficult to obtain structural data as they appear faint and blurry in images, or their disordered regions are missing altogether.
Among these techniques, NMR spectroscopy stands out for its sensitivity to protein dynamics. By combining NMR with other experimental methods, researchers can gain valuable insights into protein mobility and internal interactions.
Harnessing the power of NMR
Employing an integrated approach and combining various experimental and computational techniques helps researchers gain a comprehensive understanding of how protein structures impact their biological functions. Solid-state nuclear magnetic resonance (NMR) spectroscopy serves as the primary analytical tool in this multifaceted approach. It offers atomic-resolution information even for challenging proteins and, unlike destructive techniques, solid-state NMR harnesses the inherent magnetic properties of atomic nuclei, providing insights into structure and dynamics across diverse biological systems.
Solid-state nuclear magnetic resonance (NMR) spectroscopy serves as the primary analytical tool in this multifaceted approach. Unlike destructive techniques, it harnesses the inherent magnetic properties of atomic nuclei, providing insights into structure and dynamics across diverse biological systems.
Solid-state NMR operates by rapidly spinning the sample around a particular angle – the “magic angle” – to give magic angle spinning (MAS) that achieves high spectral resolution for non-liquid samples. Using solid-state NMR, researchers can study not only static structures, but also conformational dynamics. This unique capability allows solid-state NMR to uncover structural details hidden by other techniques, particularly in intrinsically disordered proteins and during real-time biochemical reactions.
Application 1: Unlocking the secrets of HIV maturation
Human immunodeficiency virus (HIV), the causative agent of acquired immune deficiency syndrome (AIDS), has claimed over 40 million lives globally (4). It necessitates continuous research for effective treatments to prevent HIV from progressing to AIDS. Advancements in understanding the structural basis of HIV-1 maturation and its inhibition by maturation inhibitors, such as Bevirimat (BVM), offer promising avenues for developing more potent inhibitors that could restrict HIV maturation and prevent the virus from turning infectious.
The molecular dynamics during HIV maturation present unique opportunities to interfere with HIV infection (5). As the HIV-1 virus matures, sequential cleavage of the Gag precursor by the virus protease, during the process of virus assembly (6), is vital for infectivity. The final step involves the 14-residue spacer peptide 1 (SP1) cleaved off, allowing the infectious capsids to form from the capsid protein (CA) (7). Studies have explored the potential of arresting this final step of maturation for the design of potent anti-HIV inhibitors.
Using a combination of solid-state NMR, cryo-EM and computational simulations, researchers demonstrated the critical role of SP1 peptide mobility in the cleavage process. This mobility is essential for effective cleavage requiring access of the peptide by the viral protease enzyme.
This finding sheds light on the mechanism of action of small-molecule inhibitors like BVM. BVM binds within a pore formed by six SP1 peptides, essentially immobilizing them and halting the maturation process (8). By harnessing this knowledge, scientists are developing improved maturation inhibitors, representing a novel and potentially life-saving class of drugs for combating HIV/AIDS.
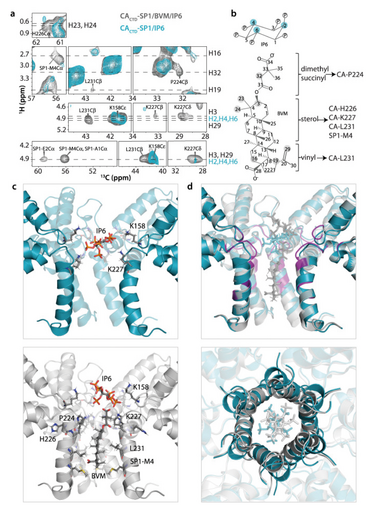
Figure 1. Solid-state NMR spectra (top) and structure of Bevirimat (BVM)- and inositol hexakisphosphate (IP6)-bound CACTD-SP1 crystalline assemblies (bottom). The chemical structures of assembly cofactor IP6 and maturation inhibitor BVM are shown next to the spectra.
Application 2: Identifying protein biomarkers for cancer
Another area of research concerns cytoskeletal protein assemblies and their potential implications for cancer biomarker discovery. This domain focuses on proteins that interact with two essential structural elements within the cell – microtubules and actin.
Microtubules, filamentous structures making up the cytoskeleton, play pivotal roles in numerous cellular processes, including maintaining cellular architecture, facilitating organelle transport, enabling cell motility and driving cell division. For these functions to be executed, microtubules need to recruit microtubule-associated proteins (MAPs).
MAPs exert significant regulatory control over microtubule formation, stability and interactions with both the cytoskeleton and other cellular components (9). Deciphering the relationship between MAP structure and function, particularly at the atomic level, holds immense promise for advancing the understanding of disease processes and paving the way for the development of novel therapeutic interventions.
Leveraging solid-state NMR, researchers have successfully determined the structure of several MAPs bound to polymeric microtubules. This includes the motor domain of kinesin-1 (10) and TPX2, a protein previously resistant to analysis due to its inherent structural disorder (11). Interruption of the TPX2 function or expression can lead to genomic instability and affect important microtubule cell division processes (12). This potential association suggests that TPX2 could serve as a valuable biomarker for malignancies, potentially enabling earlier cancer detection and facilitating the development of more effective treatment strategies.
Application 3: Exploring the role of the cytoskeleton in cellular function
Within the dynamic network of the cytoskeleton, actin filaments play a crucial role in diverse cellular functions, their polymerization meticulously regulated by actin-binding proteins. Among these regulators, the cofilin family of proteins stands out for its ability to sever actin and disassemble filaments. Despite its known function, the structural basis for this activity remained elusive, primarily due to the limitations of traditional techniques in capturing atomic-level details of cofilin-actin complexes (13).
This lack of detailed understanding is particularly concerning as disrupted actin dynamics have been implicated in various pathologies, including neurodegenerative diseases, multiple cancers, and cardiomyopathy. Unraveling the molecular mechanisms behind these disruptions is vital for developing potential therapeutic interventions.
Solid-state NMR permits the high-resolution visualization of actin-associated proteins bound to actin filaments at the atomic level, providing unprecedented insights into their interaction and regulatory mechanisms. While previous studies using cryo-EM have shed light on changes in actin filament architecture induced by cofilin, solid-state NMR uncovers the structural details of the cofilin-actin complex with atomic precision.
A final thought: Investigating proteins within living cells
There are some challenges with high-specification NMR instruments – they require housing in dedicated facilities, with teams of specialist technicians in order to operate them, for example. Additionally, the infrastructure and maintenance needs of high-field NMR can limit adoption of NMR techniques. However, ongoing efforts to increase field strength using permanent magnet technology, such as benchtop instrumentation, that requires no specialist installation or a cryogen – or water – cooled magnet can minimize the cost of ownership and make it more widely accessible. Improvements in sensitivity and resolution and further integration of NMR with other techniques can expand the scope of its analytical capabilities, increasing the viability of its use in different fields of study.
Nevertheless, the techniques established for studying isolated proteins and protein assemblies now pave the way for in situ structural investigations, pushing the boundaries of sensitivity and expanding the capabilities of MAS NMR. This exciting advancement allows researchers to directly observe the structure and dynamics of proteins within the complex environment of living cells, providing crucial insights into their natural function.
Analyzing proteins in living cells, using the non-destructive technique of NMR, can help investigate how proteins interact with drugs or inhibitors that could be used as pharmaceutical treatments, which would have beneficial implications for healthcare programs worldwide.
- A Schuhmacher et al, “Analysis of pharma R&D productivity - A new perspective needed,” Drug Discov Today, 28, 103726 (2023). DOI: 10.1016/j.drudis.2023.103726. Epub 2023 Jul 26. PMID: 37506762.
- T Polenova T, Quinn CM, Gronenborn AM (eds). “Integrated Structural Biology”. Royal Society of Chemistry (2023).
- EYD Chua et al, “Better, faster, cheaper: Recent advances in Cryo–electron microscopy,” Annu Rev Biochem, 91, 1 (2022). DOI:10.1146/annurev-biochem-032620-110705.
- Global HIV & AIDS statistics – Fact sheet, UNAIDS, 2023. https://www.unaids.org/en/resources/fact-sheet (Accessed 5 January 2024).
- M Wang et al, "Quenching protein dynamics interferes with HIV capsid maturation." Nat Commun, 8, 1779 (2017).
- P Spearman, “HIV-1 Gag as an Antiviral Target: Development of Assembly and Maturation Inhibitors,” Curr Top Med Chem, 16, 1154 (2016). DOI:10.2174/1568026615666150902102143.
- A de Marco et al, “Structural analysis of HIV-1 maturation using cryo-electron tomography,” PLoS Pathogens, 6(11): e1001215 (2010).
- S Sarkar et al, “Structural basis of HIV-1 maturation inhibitor binding and activity,” Nat Commun, 14, 1237 (2023).
- C Janke and MM Magiera, “The tubulin code and its role in controlling microtubule properties and functions,” Nat Rev Mol Cell Biol, 21, 307 (2020).
- C Zhang et al, "Magic-angle-spinning NMR structure of the kinesin-1 motor domain assembled with microtubules reveals the elusive neck linker orientation," Nat Commun, 13, 6795 (2022).
- C Guo et al, "Structural basis of protein condensation on microtubules underlying branching microtubule nucleation," Nat Commun, 14, 3682 (2023).
- I Pérez de Castro and M Malumbres, “Mitotic Stress and Chromosomal Instability in Cancer: The Case for TPX2,” Genes Cancer, 3, 721 (2012). DOI:10.1177/1947601912473306.
- J Kraus et al, "Magic angle spinning NMR structure of human cofilin-2 assembled on actin filaments reveals isoform-specific conformation and binding mode," Nat Commun, 13, 2114 (2022).
Tatyana Polenova is Professor of Chemistry and Biochemistry at the University of Delaware, Newark, USA. Her research focuses on investigating the structure and dynamics of biological systems and complex formulations in the solid state using NMR spectroscopy, combined with computational, and biochemical methods. Polenova serves as Co-Director of the Pittsburgh Center for HIV Protein Interactions.